化工进展 ›› 2025, Vol. 44 ›› Issue (5): 2451-2462.DOI: 10.16085/j.issn.1000-6613.2024-1841
• 合成生物制造 • 上一篇
甲基杆菌有效同化工业甲醇和植物甲醇的研究进展
姚陆1,2,3( ), 马增新1,2,3(
), 马增新1,2,3( ), 张聪1,2,3, 杨松1,2,3(
), 张聪1,2,3, 杨松1,2,3( ), 邢新会4,5(
), 邢新会4,5( )
)
- 1.青岛农业大学生命科学学院,山东 青岛 266109
2.青岛农业大学山东省应用真菌重点实验室,山东 青岛 266109
3.青岛农业大学青岛生物沼气环境微生物国际合作基地,山东 青岛 266109
4.清华大学工业生物催化 教育部重点实验室,化工系生物化工研究所,合成与系统生物学中心,北京 100084
5.清华大学深圳国际 研究生院生物医药与健康工程研究院,广东 深圳 518055
-
收稿日期:2024-11-11修回日期:2025-02-10出版日期:2025-05-25发布日期:2025-05-20 -
通讯作者:杨松,邢新会 -
作者简介:姚陆(1990—),男,博士,研究方向为代谢工程和微生物菌剂。E-mail:yaolu113@163.com
马增新(1988—),男,博士,研究方向为代谢工程和合成生物学。E-mail:mazengxin@foxmail.com。 -
基金资助:国家重点研发计划(2021YFC2103500);国家自然科学基金(22078169);国家自然科学基金青年基金(31900004)
Recent advances in strengthening Methylobacterium chassis for the utilization of methanol from industrial and plant sources
YAO Lu1,2,3( ), MA Zengxin1,2,3(
), MA Zengxin1,2,3( ), ZHANG Cong1,2,3, YANG Song1,2,3(
), ZHANG Cong1,2,3, YANG Song1,2,3( ), XING Xinhui4,5(
), XING Xinhui4,5( )
)
- 1.School of Life Sciences, Qingdao Agricultural University, Qingdao 266109, Shandong, China
2.Shandong Province Key Laboratory of Applied Mycology, Qingdao Agricultural University, Qingdao 266109, Shandong, China
3.Qingdao International Center on Microbes Utilizing Biogas, Qingdao Agricultural University, Qingdao 266109, Shandong, China
4.MOE Key Laboratory for Industrial Biocatalysis, Institute of Biochemical Engineering, Department of Chemical Engineering, Center for Synthetic & Systems Biology, Tsinghua University, Beijing 100084, China
5.Institute of Biopharmaceutical and Health Engineering, Tsinghua Shenzhen International Graduate School, Shenzhen 518055, Guangdong, China
-
Received:2024-11-11Revised:2025-02-10Online:2025-05-25Published:2025-05-20 -
Contact:YANG Song, XING Xinhui
摘要:
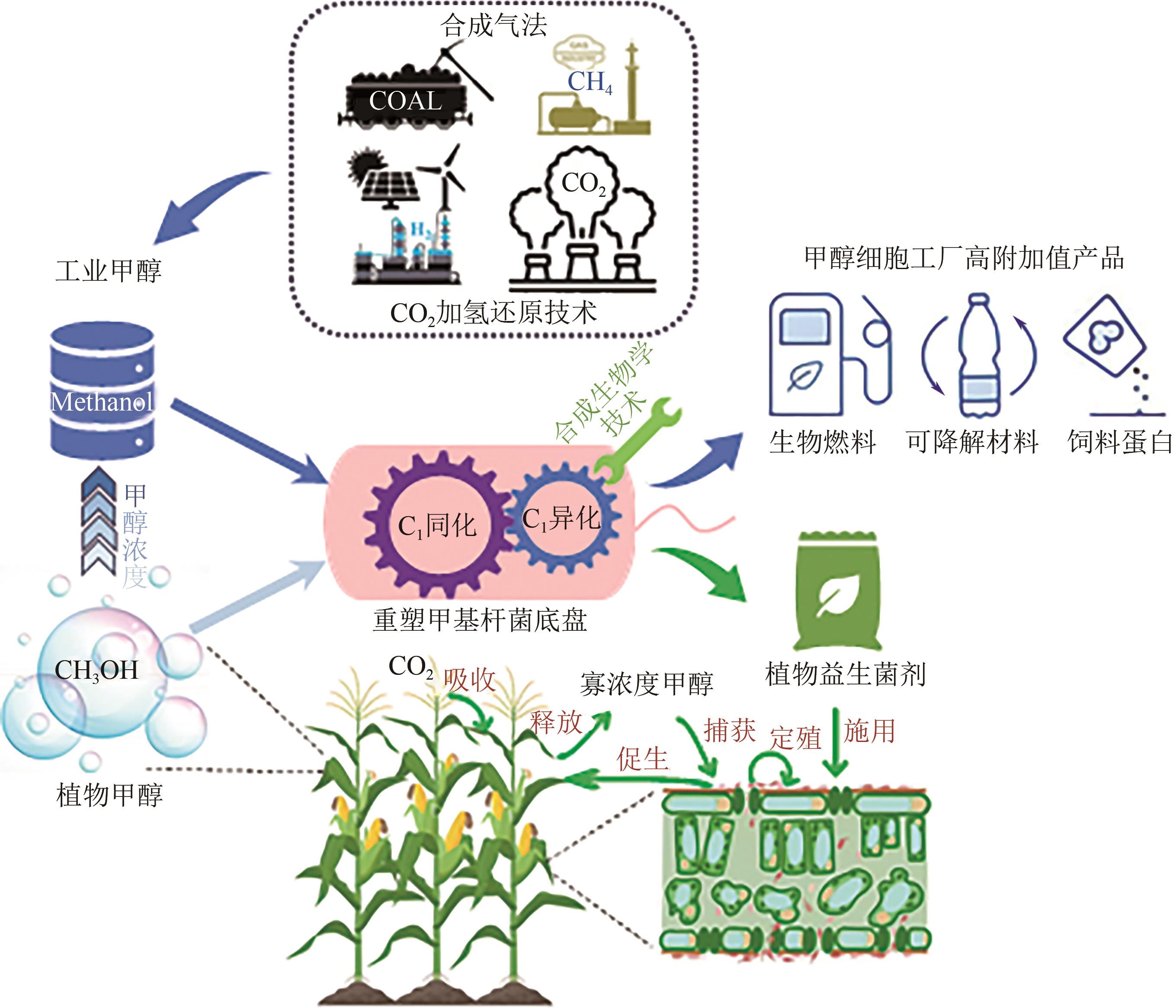
甲醇是第三代工业生物制造的重要原料,同时又能伴随植物生长排放到大气中加剧温室效应。甲基杆菌是一类能以甲醇作为生长碳源和能源的极具广泛应用前景的菌株。然而甲基杆菌催化转化高浓度工业甲醇和寡浓度植物甲醇的过程效率偏低,导致甲醇的碳和电子耗散,制约了甲醇的高值化利用。本文首先系统综述了甲基杆菌底盘再造强化工业甲醇有效代谢方面的最新策略,进而论述了甲基杆菌对植物源寡浓度甲醇有效氧化、同化和转化的最新认知,最后展望了甲基杆菌进一步强化甲醇利用的可行性研究方向与策略。本综述期望推动甲基杆菌作为甲醇基细胞工厂和新型植物益生菌剂的技术创新,助力工业生物制造的甲醇高值化,增强植物甲醇的固定封存,实现经济与生态环境效益的共赢发展。
中图分类号:
引用本文
姚陆, 马增新, 张聪, 杨松, 邢新会. 甲基杆菌有效同化工业甲醇和植物甲醇的研究进展[J]. 化工进展, 2025, 44(5): 2451-2462.
YAO Lu, MA Zengxin, ZHANG Cong, YANG Song, XING Xinhui. Recent advances in strengthening Methylobacterium chassis for the utilization of methanol from industrial and plant sources[J]. Chemical Industry and Engineering Progress, 2025, 44(5): 2451-2462.
| 产品 | 代谢工程策略 | 发酵规模 | 产量 | 参考文献 |
|---|---|---|---|---|
| 单细胞蛋白 | 高密度发酵 | 发酵罐 | 42~72g/L | [ |
| 聚β-羟基丁酸酯 | 限氮两阶段发酵 | 发酵罐 | 46~136g/L | [ |
| 聚β-羟基丁酸-戊酸-己酸酯 | 替换本源phaC,过表达β-酮硫解酶和乙酰乙酰辅酶A还原酶 | 摇瓶 | 43.6% | [ |
| 衣康酸 | 表达顺乌头酸脱羧酶,并解除PhaR调控 | 摇瓶 | 31.6mg/L | [ |
| 异丁醇 | 优化异丁醇合成途径基因及拷贝,减少竞争途径乳酸合成 | 摇瓶 | 19mg/L | [ |
| 3-羟基丙酸 | 表达mcr并优化启动子和拷贝数 | 摇瓶 | 69.8mg/L | [ |
| 3-羟基丙酸 | 构建协同同化途径,强化ATP和还原力供给 | 发酵罐 | 0.86g/L | [ |
| 3-羟基丙酸 | 构建光合甲基营养型菌株,强化能量供给 | 发酵罐 | 1.8g/L | [ |
| 蛇麻烯 | 表达甲羟戊酸途径和蛇麻烯合酶 | 发酵罐 | 1.65g/L | [ |
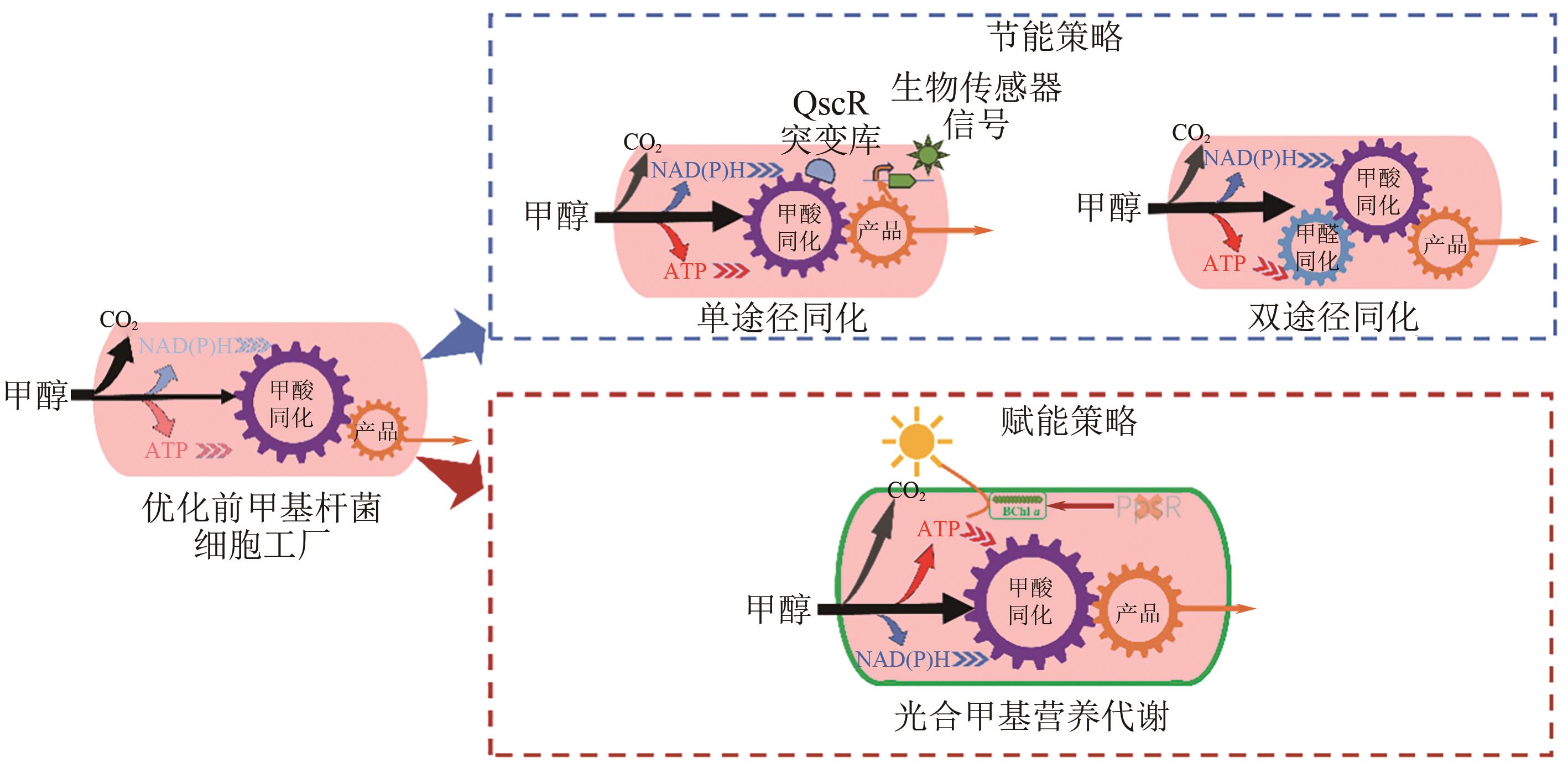
| 甲羟戊酸 | 构建甲羟戊酸生物传感器对转录因子QscR进行筛选 | 发酵罐 | 2.67g/L | [ |
| 筛选3-羟基丁酰辅酶A变位酶并进行限氮两阶段发酵 | 发酵罐 | 2.1g/L | [ | |
| 类胡萝卜素 | 利用CRISPRi打靶竞争途径 | 摇瓶 | 0.3μg/mg | [ |
| 紫色杆菌素 | 表达紫色杆菌素基因簇vioABCDE并进行随机突变筛选 | 摇瓶 | 118mg/L | [ |
表1 扭脱甲基杆菌合成大宗及精细化学品
| 产品 | 代谢工程策略 | 发酵规模 | 产量 | 参考文献 |
|---|---|---|---|---|
| 单细胞蛋白 | 高密度发酵 | 发酵罐 | 42~72g/L | [ |
| 聚β-羟基丁酸酯 | 限氮两阶段发酵 | 发酵罐 | 46~136g/L | [ |
| 聚β-羟基丁酸-戊酸-己酸酯 | 替换本源phaC,过表达β-酮硫解酶和乙酰乙酰辅酶A还原酶 | 摇瓶 | 43.6% | [ |
| 衣康酸 | 表达顺乌头酸脱羧酶,并解除PhaR调控 | 摇瓶 | 31.6mg/L | [ |
| 异丁醇 | 优化异丁醇合成途径基因及拷贝,减少竞争途径乳酸合成 | 摇瓶 | 19mg/L | [ |
| 3-羟基丙酸 | 表达mcr并优化启动子和拷贝数 | 摇瓶 | 69.8mg/L | [ |
| 3-羟基丙酸 | 构建协同同化途径,强化ATP和还原力供给 | 发酵罐 | 0.86g/L | [ |
| 3-羟基丙酸 | 构建光合甲基营养型菌株,强化能量供给 | 发酵罐 | 1.8g/L | [ |
| 蛇麻烯 | 表达甲羟戊酸途径和蛇麻烯合酶 | 发酵罐 | 1.65g/L | [ |
| 甲羟戊酸 | 构建甲羟戊酸生物传感器对转录因子QscR进行筛选 | 发酵罐 | 2.67g/L | [ |
| 筛选3-羟基丁酰辅酶A变位酶并进行限氮两阶段发酵 | 发酵罐 | 2.1g/L | [ | |
| 类胡萝卜素 | 利用CRISPRi打靶竞争途径 | 摇瓶 | 0.3μg/mg | [ |
| 紫色杆菌素 | 表达紫色杆菌素基因簇vioABCDE并进行随机突变筛选 | 摇瓶 | 118mg/L | [ |
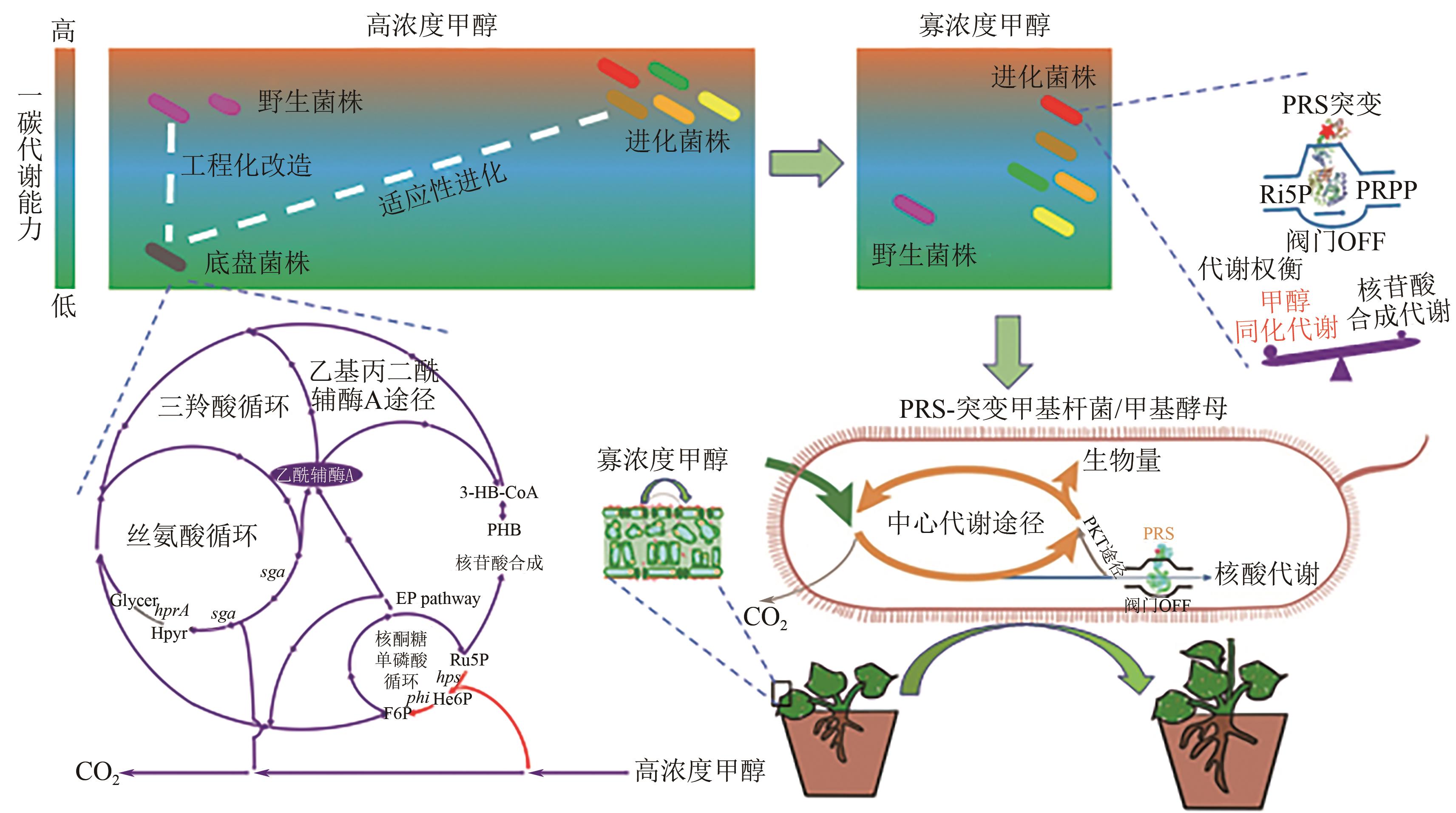
图3 甲基杆菌代谢阀调控植物寡浓度甲醇高效利用的同时促农作物生长Ru5P—核酮糖-5-磷酸;F6P—果糖-6-磷酸;PHB—聚β-羟基丁酸;He6P—己酮糖-6-磷酸;Glycer—甘油酸;Hypr—羟基丙酮酸;Ri5P—核糖-5-磷酸;PRPP—磷酸核糖焦磷酸;PRS—磷酸核糖焦磷酸合成酶;PKT pathway—磷酸转酮酶途径
| 1 | GUO Feng, QIAO Yangyi, XIN Fengxue, et al. Bioconversion of C1 feedstocks for chemical production using Pichia pastoris[J]. Trends in Biotechnology, 2023, 41(8): 1066-1079. |
| 2 | 姚伦, 周雍进. 一碳化合物生物利用和转化研究进展[J]. 化工进展, 2023, 42(1): 16-29. |
| YAO Lun, ZHOU Yongjin. Progress in microbial utilization of one-carbon feedstocks for biomanufacturing[J]. Chemical Industry and Engineering Progress, 2023, 42(1): 16-29. | |
| 3 | 李灿. 绿色氢能在“双碳” 战略中的作用[J]. 科技导报, 2024, 42(15): 1-2. |
| LI Can. The role of green hydrogen energy in dual carbon strategy[J]. Science & Technology Review, 2024, 42(15): 1-2. | |
| 4 | CAI Peng, WU Xiaoyan, DENG Jun, et al. Methanol biotransformation toward high-level production of fatty acid derivatives by engineering the industrial yeast Pichia pastoris [J]. Proceedings of the National Academy of Sciences of the United States of America, 2022, 119(29): e2201711119. |
| 5 | GUO Yuanke, ZHANG Rui, WANG Jing, et al. Engineering yeasts to co-utilize methanol or formate coupled with CO2 fixation[J]. Metabolic Engineering, 2024, 84: 1-12. |
| 6 | LIU Zihe, WANG Kai, CHEN Yun, et al. Third-generation biorefineries as the means to produce fuels and chemicals from CO2 [J]. Nature Catalysis, 2020, 3: 274-288. |
| 7 | MILLET D B, JACOB D J, CUSTER T G, et al. New constraints on terrestrial and oceanic sources of atmospheric methanol[J]. Atmospheric Chemistry & Physics, 2008, 8(149): 6887-6905. |
| 8 | KOLB Steffen. Aerobic methanol-oxidizing bacteria in soil[J]. FEMS Microbiology Letters, 2009, 300(1): 1-10. |
| 9 | ZHANG Min, YUAN Xiaojie, ZHANG Cong, et al. Bioconversion of methanol into value-added chemicals in native and synthetic methylotrophs[J]. Current Issues in Molecular Biology, 2019, 33: 225-236. |
| 10 | CUI Lanyu, YANG Jing, LIANG Weifan, et al. Sodium formate redirects carbon flux and enhances heterologous mevalonate production in Methylobacterium extorquens AM1[J]. Biotechnology Journal, 2023, 18(2): e2200402. |
| 11 | MA Zengxin, ZHANG Min, ZHANG Changtai, et al. Metabolomic analysis improves bioconversion of methanol to isobutanol in Methylorubrum extorquens AM1[J]. Biotechnology Journal, 2021, 16(6): e2000413. |
| 12 | YOON Jihee, Jiyun BAE, KANG Seulgi, et al. Poly-3-hydroxybutyrate production in acetate minimal medium using engineered Methylorubrum extorquens AM1[J]. Bioresource Technology, 2022, 353: 127127. |
| 13 | HURT Allison, BIBIK Jacob D, MARTINEZ-GOMEZ Norma Cecilia, et al. Engineering terpene production pathways in Methylobacterium extorquens AM1[J]. Microorganisms, 2024, 12(3): 500. |
| 14 | SALZE Guillaume P, TIBBETTS Sean M. Apparent digestibility coefficients of proximate nutrients and essential amino acids from a single-cell protein meal derived from Methylobacterium extorquens for pre-smolt Atlantic salmon (Salmo salar L.)[J]. Aquaculture Research, 2021, 52(12): 6818-6823. |
| 15 | JONES Shawn W, KARPOL Alon, FRIEDMAN Sivan, et al. Recent advances in single cell protein use as a feed ingredient in aquaculture[J]. Current Opinion in Biotechnology, 2020, 61: 189-197. |
| 16 | BÉLANGER L, FIGUEIRA M M, BOURQUE D, et al. Production of heterologous protein by Methylobacterium extorquens in high cell density fermentation[J]. FEMS Microbiology Letters, 2004, 231(2): 197-204. |
| 17 | TLUSTY Michael, RHYNE Andrew, SZCZEBAK Joseph T, et al. A transdisciplinary approach to the initial validation of a single cell protein as an alternative protein source for use in aquafeeds[J]. PeerJ, 2017, 5: e3170. |
| 18 | SUZUKI Takahiro, YAMANE Tsuneo, SHIMIZU Shoichi. Mass production of poly-β-hydroxybutyric acid by fully automatic fed-batch culture of methylotroph[J]. Applied Microbiology and Biotechnology, 1986, 23(5): 322-329. |
| 19 | BOURQUE D, POMERLEAU Y, GROLEAU D. High-cell-density production of poly-β-hydroxybutyrate (PHB) from methanol by Methylobacterium extorquens: Production of high-molecular-mass PHB[J]. Applied Microbiology and Biotechnology, 1995, 44(3): 367-376. |
| 20 | ORITA Izumi, NISHIKAWA Kouta, NAKAMURA Satoshi, et al. Biosynthesis of polyhydroxyalkanoate copolymers from methanol by Methylobacterium extorquens AM1 and the engineered strains under cobalt-deficient conditions[J]. Applied Microbiology and Biotechnology, 2014, 98(8): 3715-3725. |
| 21 | Chee Kent LIM, VILLADA Juan C, CHALIFOUR Annie, et al. Designing and engineering Methylorubrum extorquens AM1 for itaconic acid production[J]. Frontiers in Microbiology, 2019, 10: 1027. |
| 22 | YANG Yiming, CHEN Wenjing, YANG Jing, et al. Production of 3-hydroxypropionic acid in engineered Methylobacterium extorquens AM1 and its reassimilation through a reductive route[J]. Microbial Cell Factories, 2017, 16(1): 179. |
| 23 | YUAN Xiaojie, CHEN Wenjing, MA Zengxin, et al. Rewiring the native methanol assimilation metabolism by incorporating the heterologous ribulose monophosphate cycle into Methylorubrum extorquens [J]. Metabolic Engineering, 2021, 64: 95-110. |
| 24 | MA Zengxin, FENG Chenxi, SONG Yazhen, et al. Engineering photo-methylotrophic Methylobacterium for enhanced 3-hydroxypropionic acid production during non-growth stage fermentation[J]. Bioresource Technology, 2024, 393: 130104. |
| 25 | SONNTAG Frank, KRONER Cora, LUBUTA Patrice, et al. Engineering Methylobacterium extorquens for de novo synthesis of the sesquiterpenoid α-humulene from methanol[J]. Metabolic Engineering, 2015, 32: 82-94. |
| 26 | LIANG Weifan, CUI Lanyu, CUI Jinyu, et al. Biosensor-assisted transcriptional regulator engineering for Methylobacterium extorquens AM1 to improve mevalonate synthesis by increasing the acetyl-CoA supply[J]. Metabolic Engineering, 2017, 39: 159-168. |
| 27 | ROHDE Maria-Teresa, TISCHER Sylvi, HARMS Hauke, et al. Production of 2-hydroxyisobutyric acid from methanol by Methylobacterium extorquens AM1 expressing (R)-3-hydroxybutyryl coenzyme A-isomerizing enzymes[J]. Applied and Environmental Microbiology, 2017, 83(3): e02622-16. |
| 28 | MO Xuhua, ZHANG Hui, WANG Tianmin, et al. Establishment of CRISPR interference in Methylorubrum extorquens and application of rapidly mining a new phytoene desaturase involved in carotenoid biosynthesis[J]. Applied Microbiology and Biotechnology, 2020, 104(10): 4515-4532. |
| 29 | QUYNH LE Hoa Thi, Dung Hoang ANH MAI, NA Jeong-Geol, et al. Development of Methylorubrum extorquens AM1 as a promising platform strain for enhanced violacein production from co-utilization of methanol and acetate[J]. Metabolic Engineering, 2022, 72: 150-160. |
| 30 | ZHANG Cong, ZHOU Difei, WANG Mengying, et al. Phosphoribosylpyrophosphate synthetase as a metabolic valve advances Methylobacterium/Methylorubrum phyllosphere colonization and plant growth[J]. Nature Communications, 2024, 15(1): 5969. |
| 31 | VERA Rocío Torres, GARCÍA Antonio José Bernabé, ÁLVAREZ Francisco José Carmona, et al. Application and effectiveness of Methylobacterium symbioticum as a biological inoculant in maize and strawberry crops[J]. Folia Microbiologica, 2024, 69(1): 121-131. |
| 32 | GROSSI Cecilia E M, TANI Akio, MORI Izumi C, et al. Plant growth-promoting abilities of Methylobacterium sp. 2A involve auxin-mediated regulation of the root architecture[J]. Plant, Cell & Environment, 2024, 47(12): 5343-5357. |
| 33 | VADIVUKKARASI Ponnusamy, SUSEELA BHAI R. Phyllosphere-associated Methylobacterium: A potential biostimulant for ginger (Zingiber officinale Rosc.) cultivation[J]. Archives of Microbiology, 2020, 202(2): 369-375. |
| 34 | SENTHILKUMAR M, PUSHPAKANTH P, Arul JOSE P, et al. Diversity and functional characterization of endophytic Methylobacterium isolated from banana cultivars of South India and its impact on early growth of tissue culture banana plantlets[J]. Journal of Applied Microbiology, 2021, 131(5): 2448-2465. |
| 35 | ZHU Liping, SONG Yazhen, MA Shunan, et al. Heterologous production of 3-hydroxypropionic acid in Methylorubrum extorquens by introducing the MCR gene via a multi-round chromosomal integration system based on cre-lox71/lox66 and transposon[J]. Microbial Cell Factories, 2024, 23(1): 5. |
| 36 | ZHU Wenliang, CUI Jinyu, CUI Lanyu, et al. Bioconversion of methanol to value-added mevalonate by engineered Methylobacterium extorquens AM1 containing an optimized mevalonate pathway[J]. Applied Microbiology and Biotechnology, 2016, 100(5): 2171-2182. |
| 37 | PEYRAUD Rémi, SCHNEIDER Kathrin, KIEFER Patrick, et al. Genome-scale reconstruction and system level investigation of the metabolic network of Methylobacterium extorquens AM1[J]. BMC Systems Biology, 2011, 5: 189. |
| 38 | YANG Jing, ZHANG Changtai, YUAN Xiaojie, et al. Metabolic engineering of Methylobacterium extorquens AM1 for the production of butadiene precursor[J]. Microbial Cell Factories, 2018, 17(1): 194. |
| 39 | KRÜSEMANN Jan L, RAINALDI Vittorio, COTTON Charles AR, et al. The cofactor challenge in synthetic methylotrophy: Bioengineering and industrial applications[J]. Current Opinion in Biotechnology, 2023, 82: 102953. |
| 40 | ZHAN Chunjun, LI Xiaowei, YANG Yankun, et al. Strategies and challenges with the microbial conversion of methanol to high-value chemicals[J]. Biotechnology and Bioengineering, 2021, 118(10): 3655-3668. |
| 41 | SINGH Hawaibam Birla, KANG Min-Kyoung, KWON Moonhyuk, et al. Developing methylotrophic microbial platforms for a methanol-based bioindustry[J]. Frontiers in Bioengineering and Biotechnology, 2022, 10: 1050740. |
| 42 | CUI Jinyu, GOOD Nathan M, HU Bo, et al. Metabolomics revealed an association of metabolite changes and defective growth in Methylobacterium extorquens AM1 overexpressing ecm during growth on methanol[J]. PLoS One, 2016, 11(4): e0154043. |
| 43 | DENG Chen, WU Yaokang, Xueqin Lyu, et al. Refactoring transcription factors for metabolic engineering[J]. Biotechnology Advances, 2022, 57: 107935. |
| 44 | KALYUZHNAYA Marina G, LIDSTROM Mary E. QscR-mediated transcriptional activation of serine cycle genes in Methylobacterium extorquens AM1[J]. Journal of Bacteriology, 2005, 187(21): 7511-7517. |
| 45 | YU Hong, LIAO James C. A modified serine cycle in Escherichia coli coverts methanol and CO2 to two-carbon compounds[J]. Nature Communications, 2018, 9(1): 3992. |
| 46 | HE Hai, Rune HÖPER, Moritz DODENHÖFT, et al. An optimized methanol assimilation pathway relying on promiscuous formaldehyde-condensing aldolases in E. coli [J]. Metabolic Engineering, 2020, 60: 1-13. |
| 47 | VON BORZYSKOWSKI Lennart Schada, SONNTAG Frank, Laura PÖSCHEL, et al. Replacing the ethylmalonyl-CoA pathway with the glyoxylate shunt provides metabolic flexibility in the central carbon metabolism of Methylobacterium extorquens AM1[J]. ACS Synthetic Biology, 2018, 7(1): 86-97. |
| 48 | NIEH Liang-Yu, CHEN Frederic Y-H, JUNG Hsin-Wei, et al. Evolutionary engineering of methylotrophic E. coli enables fast growth on methanol[J]. Nature Communications, 2024, 15(1): 8840. |
| 49 | REITER Michael A, BRADLEY Timothy, BÜCHEL Lars A, et al. A synthetic methylotrophic Escherichia coli as a chassis for bioproduction from methanol[J]. Nature Catalysis, 2024, 7(5): 560-573. |
| 50 | WANG Guokun, Mattis OLOFSSON-DOLK, HANSSON Frederik Gleerup, et al. Engineering yeast Yarrowia lipolytica for methanol assimilation[J]. ACS Synthetic Biology, 2021, 10(12): 3537-3550. |
| 51 | WANG Weiting, GUO Shuqi, HOU Qianzi, et al. Bioupcycling methane and CO2 for succinate production by an engineered type Ⅰ methanotrophic bacterium[J]. Journal of Agricultural and Food Chemistry, 2024, 72(44): 24237-24245. |
| 52 | ZHAN Chunjun, LI Xiaowei, LAN Guangxu, et al. Reprogramming methanol utilization pathways to convert Saccharomyces cerevisiae to a synthetic methylotroph[J]. Nature Catalysis, 2023, 6(5): 435-450. |
| 53 | PETERSON Autumn, BASKETT Carina, RATCLIFF William C, et al. Transforming yeast into a facultative photoheterotroph via expression of vacuolar rhodopsin[J]. Current Biology, 2024, 34(3): 648-654.e3. |
| 54 | HU Guipeng, LI Zehong, MA Danlei, et al. Light-driven CO2 sequestration in Escherichia coli to achieve theoretical yield of chemicals[J]. Nature Catalysis, 2021, 4: 395-406. |
| 55 | DAHLIN Lukas R, MEYERS Alex W, STEFANI Skylar W, et al. Heterologous expression of formate dehydrogenase enables photoformatotrophy in the emerging model microalga, Picochlorum renovo [J]. Frontiers in Bioengineering and Biotechnology, 2023, 11: 1162745. |
| 56 | ALESSA Ola, OGURA Yoshitoshi, FUJITANI Yoshiko, et al. Comprehensive comparative genomics and phenotyping of Methylobacterium species[J]. Frontiers in Microbiology, 2021, 12: 740610. |
| 57 | KUZYK Steven B, MESSNER Katia, PLOUFFE Jocelyn, et al. Diverse aerobic anoxygenic phototrophs synthesize bacteriochlorophyll in oligotrophic rather than copiotrophic conditions, suggesting ecological niche[J]. Environmental Microbiology, 2023, 25(11): 2653-2665. |
| 58 | STIEFEL Philipp, ZAMBELLI Tomaso, VORHOLT Julia A. Isolation of optically targeted single bacteria by application of fluidic force microscopy to aerobic anoxygenic phototrophs from the phyllosphere[J]. Applied and Environmental Microbiology, 2013, 79(16): 4895-4905. |
| 59 | YURIMOTO Hiroya, SAKAI Yasuyoshi. Interaction between C1-microorganisms and plants: Contribution to the global carbon cycle and microbial survival strategies in the phyllosphere[J]. Bioscience, Biotechnology, and Biochemistry, 2022, 87(1): 1-6. |
| 60 | YURIMOTO Hiroya, SHIRAISHI Kosuke, SAKAI Yasuyoshi. Physiology of methylotrophs living in the phyllosphere[J]. Microorganisms, 2021, 9(4): 809. |
| 61 | GOURION Benjamin, ROSSIGNOL Michel, VORHOLT Julia A. A proteomic study of Methylobacterium extorquens reveals a response regulator essential for epiphytic growth[J]. Proceedings of the National Academy of Sciences of the United States of America, 2006, 103(35): 13186-13191. |
| 62 | TORGONSKAYA M L, FIRSOVA Y E, EKIMOVA G A, et al. Distribution and potential ecophysiological roles of multiple GroEL chaperonins in pink-pigmented facultative methylotrophs[J]. Microbiology, 2024, 93(1): 14-27. |
| 63 | IGUCHI Hiroyuki, YOSHIDA Yusuke, FUJISAWA Kento, et al. KaiC family proteins integratively control temperature-dependent UV resistance in Methylobacterium extorquens AM1[J]. Environmental Microbiology Reports, 2018, 10(6): 634-643. |
| 64 | TOLA Yosef Hamba, FUJITANI Yoshiko, TANI Akio. Bacteria with natural chemotaxis towards methanol revealed by chemotaxis fishing technique[J]. Bioscience, Biotechnology, and Biochemistry, 2019, 83(11): 2163-2171. |
| 65 | TANI Akio, MASUDA Sachiko, FUJITANI Yoshiko, et al. Metabolism-linked methylotaxis sensors responsible for plant colonization in Methylobacterium aquaticum strain 22A[J]. Frontiers in Microbiology, 2023, 14: 1258452. |
| 66 | OCHSNER Andrea M, HEMMERLE Lucas, VONDERACH Thomas, et al. Use of rare-earth elements in the phyllosphere colonizer Methylobacterium extorquens PA1[J]. Molecular Microbiology, 2019, 111(5): 1152-1166. |
| 67 | JUMA Patrick Otieno, FUJITANI Yoshiko, ALESSA Ola, et al. Siderophore for lanthanide and iron uptake for methylotrophy and plant growth promotion in Methylobacterium aquaticum strain 22A[J]. Frontiers in Microbiology, 2022, 13: 921635. |
| 68 | FEATHERSTON Emily R, COTRUVO Joseph A. The biochemistry of lanthanide acquisition, trafficking, and utilization[J]. Biochimica et Biophysica Acta (BBA): Molecular Cell Research, 2021, 1868(1): 118864. |
| 69 | GROOM Joseph D, FORD Stephanie M, PESESKY Mitchell W, et al. A mutagenic screen identifies a TonB-dependent receptor required for the lanthanide metal switch in the type Ⅰ methanotroph “Methylotuvimicrobium buryatense” 5GB1C[J]. Journal of Bacteriology, 2019, 201(15): e00120-19. |
| 70 | CHU Frances, LIDSTROM Mary E. XoxF acts as the predominant methanol dehydrogenase in the type Ⅰ methanotroph Methylomicrobium buryatense [J]. Journal of Bacteriology, 2016, 198(8): 1317-1325. |
| 71 | HE Chenyi, FENG Yiping, DENG Yirong, et al. A systematic review and meta-analysis on the root effects and toxic mechanisms of rare earth elements[J]. Chemosphere, 2024, 363: 142951. |
| 72 | JIAO Yunlong, HE Ding, ZHANG Shuya, et al. Lanthanum interferes with the fundamental rhythms of stomatal opening, expression of related genes, and evapotranspiration in Arabidopsis thaliana [J]. Ecotoxicology and Environmental Safety, 2024, 281: 116576. |
| 73 | BAZURTO Jannell V, RIAZI Siavash, Simon D’ALTON, et al. Global transcriptional response of Methylorubrum extorquens to formaldehyde stress expands the role of EfgA and is distinct from antibiotic translational inhibition[J]. Microorganisms, 2021, 9(2): 347. |
| 74 | BAZURTO Jannell V, NAYAK Dipti D, TICAK Tomislav, et al. EfgA is a conserved formaldehyde sensor that leads to bacterial growth arrest in response to elevated formaldehyde[J]. PLoS Biology, 2021, 19(5): e3001208. |
| 75 | LEE Jessica A, RIAZI Siavash, NEMATI Shahla, et al. Microbial phenotypic heterogeneity in response to a metabolic toxin: Continuous, dynamically shifting distribution of formaldehyde tolerance in Methylobacterium extorquens populations[J]. PLoS Genetics, 2019, 15(11): e1008458. |
| 76 | BAZURTO Jannell V, BRUGER Eric L, LEE Jessica A, et al. Formaldehyde-responsive proteins, TtmR and EfgA, reveal a tradeoff between formaldehyde resistance and efficient transition to methylotrophy in Methylorubrum extorquens [J]. Journal of Bacteriology, 2021, 203(9): e00589-20. |
| 77 | OCHSNER Andrea M, CHRISTEN Matthias, HEMMERLE Lucas, et al. Transposon sequencing uncovers an essential regulatory function of phosphoribulokinase for methylotrophy[J]. Current Biology, 2017, 27(17): 2579-2588.e6. |
| 78 | HENARD Calvin A, SMITH Holly K, GUARNIERI Michael T. Phosphoketolase overexpression increases biomass and lipid yield from methane in an obligate methanotrophic biocatalyst[J]. Metabolic Engineering, 2017, 41: 152-158. |
| 79 | CARLSTRÖM Charlotte I, FIELD Christopher M, Miriam BORTFELD-MILLER, et al. Synthetic microbiota reveal priority effects and keystone strains in the Arabidopsis phyllosphere[J]. Nature Ecology & Evolution, 2019, 3(10): 1445-1454. |
| 80 | HOYT Kathryn O, WOOLSTON Benjamin M. Adapting isotopic tracer and metabolic flux analysis approaches to study C1 metabolism[J]. Current Opinion in Biotechnology, 2022, 75: 102695. |
| 81 | HE Lian, FU Yanfen, LIDSTROM Mary E. Quantifying methane and methanol metabolism of “Methylotuvimicrobium buryatense” 5GB1C under substrate limitation[J]. mSystems, 2019, 4(6): 10.1128/msystems.00748-10.1128/msystems.00719. |
| 82 | SONG Yazhen, FENG Chenxi, ZHOU Difei, et al. Constructing efficient bacterial cell factories to enable one-carbon utilization based on quantitative biology: A review[J]. Quantitative Biology, 2024, 12(1): 1-14. |
| 83 | JIAN Xingjin, GUO Xiaojie, WANG Jia, et al. Microbial microdroplet culture system (MMC): An integrated platform for automated, high-throughput microbial cultivation and adaptive evolution[J]. Biotechnology and Bioengineering, 2020, 117(6): 1724-1737. |
| 84 | WANG Jia, JIAN Xingjin, XING Xinhui, et al. Empowering a methanol-dependent Escherichia coli via adaptive evolution using a high-throughput microbial microdroplet culture system[J]. Frontiers in Bioengineering and Biotechnology, 2020, 8: 570. |
| 85 | TIAN Jinzhong, DENG Wangshuying, ZHANG Ziwen, et al. Discovery and remodeling of Vibrio natriegens as a microbial platform for efficient formic acid biorefinery[J]. Nature Communications, 2023, 14(1): 7758. |
| 86 | YANG Xue, ZHANG Yanfei, ZHAO Guoping. Artificial carbon assimilation: From unnatural reactions and pathways to synthetic autotrophic systems[J]. Biotechnology Advances, 2024, 70: 108294. |
| 87 | WANG Xueying, FENG Yanbin, GUO Xiaojia, et al. Creating enzymes and self-sufficient cells for biosynthesis of the non-natural cofactor nicotinamide cytosine dinucleotide[J]. Nature Communications, 2021, 12(1): 2116. |
| 88 | BLACK William B, ZHANG Linyue, Wai Shun MAK, et al. Engineering a nicotinamide mononucleotide redox cofactor system for biocatalysis[J]. Nature Chemical Biology, 2020, 16(1): 87-94. |
| 89 | SELYANIN Vadim, HAURUSEU Dzmitry, Michal KOBLÍŽEK. The variability of light-harvesting complexes in aerobic anoxygenic phototrophs[J]. Photosynthesis Research, 2016, 128(1): 35-43. |
| 90 | CHEN Haihong, WANG Yaohong, WANG Weishan, et al. High-yield porphyrin production through metabolic engineering and biocatalysis[J]. Nature Biotechnology, 2024. |
| 91 | FOONG Lian Chee, Carmen Wai Leng LOH, Hui Suan NG, et al. Recent development in the production strategies of microbial carotenoids[J]. World Journal of Microbiology and Biotechnology, 2021, 37⑴: 12. |
| 92 | MO Xuhua, SUN Yuman, BI Yuxing, et al. Characterization of C30 carotenoid and identification of its biosynthetic gene cluster in Methylobacterium extorquens AM1[J]. Synthetic and Systems Biotechnology, 2023, 8(3): 527-535. |
| 93 | MENG Yujie, LI Shuang, ZHANG Chong, et al. Strain-level profiling with picodroplet microfluidic cultivation reveals host-specific adaption of honeybee gut symbionts[J]. Microbiome, 2022, 10(1): 140. |
| [1] | 吴孟勤, 王佳瑶, 徐友强, 王钰. 化学-生物级联转化CO2合成单细胞蛋白研究进展[J]. 化工进展, 2025, 44(5): 2429-2440. |
| [2] | 王媛媛, 张翀, 韩双艳, 邢新会. 毕赤酵母利用甲醇生产重组蛋白技术的研究进展[J]. 化工进展, 2025, 44(5): 2441-2450. |
| [3] | 高建刚, 姜亚鹏, 包宝青, 王书琦, 崔书明. 绿氢转化制绿色甲醇与绿氨[J]. 化工进展, 2025, 44(4): 1987-1997. |
| [4] | 朱国瑜, 葛棋, 付名利. 甲醇重整制氢催化剂耐久性评价和寿命预测方法[J]. 化工进展, 2025, 44(3): 1338-1346. |
| [5] | 左骥, 罗莉, 谢永锴, 陈文尧, 钱刚, 周兴贵, 段学志. 甲醇无氧脱氢制甲醛Cu催化剂的粒径效应[J]. 化工进展, 2025, 44(3): 1347-1354. |
| [6] | 韩英娜, 李丽, 张林子, 安金泽, 李文秀, 张弢. 离子液体萃取精馏分离甲醇-乙腈共沸物[J]. 化工进展, 2025, 44(2): 660-668. |
| [7] | 孙悦鹏, 孙延吉, 潘艳秋, 王成宇. 基于BO-LSTM的低温甲醇洗净化气CO2含量预测[J]. 化工进展, 2025, 44(2): 688-697. |
| [8] | 周渝, 唐甜, 熊子悠, 韦奇. 基于两级微通道分离工艺的甲醇制烯烃废水深度处理[J]. 化工进展, 2025, 44(1): 100-108. |
| [9] | 胡洋, 韩传军, 胡强, 李汶颖, 安全成, 苏洋, 武洪松, 袁果. 固体氧化物燃料电池用甲醇水蒸气重整反应器研究进展[J]. 化工进展, 2025, 44(1): 169-183. |
| [10] | 周渝, 夏太阳, 韦奇, 唐甜, 田磊. 微通道耦合反渗透膜串联处理甲醇制烯烃废水工艺优化[J]. 化工进展, 2024, 43(S1): 43-51. |
| [11] | 龙涛, 周锋, 张伟, 吴泓, 王建, 陈霖. CO-CO2体系制备氘代甲醇催化剂的合成与改性[J]. 化工进展, 2024, 43(8): 4411-4420. |
| [12] | 郭鹏, 李红伟, 李贵贤, 季东, 王东亮, 赵新红. 直接甲醇燃料电池阳极催化剂的失活机制及应对策略[J]. 化工进展, 2024, 43(7): 3812-3823. |
| [13] | 方峣, 刘雷, 高志华, 黄伟, 左志军. 光辅助直接甲醇燃料电池阳极催化剂的研究进展[J]. 化工进展, 2024, 43(5): 2611-2628. |
| [14] | 周运桃, 王洪星, 李新刚, 崔丽凤. CeO2载体在CO2加氢制甲醇中的应用和研究进展[J]. 化工进展, 2024, 43(5): 2723-2738. |
| [15] | 周秋明, 牛丛丛, 吕帅帅, 李红伟, 文富利, 徐润, 李明丰. 通过产物转化分离推动CO2加氢制甲醇过程的研究进展[J]. 化工进展, 2024, 43(5): 2776-2785. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||