化工进展 ›› 2025, Vol. 44 ›› Issue (5): 2441-2450.DOI: 10.16085/j.issn.1000-6613.2024-1843
• 合成生物制造 • 上一篇
毕赤酵母利用甲醇生产重组蛋白技术的研究进展
王媛媛1,2( ), 张翀3,4, 韩双艳5, 邢新会1,2,3,4(
), 张翀3,4, 韩双艳5, 邢新会1,2,3,4( )
)
- 1.清华大学深圳国际研究生院,生物医药与健康工程研究院,广东 深圳 518055
2.清华大学深圳国际研究生院,活性蛋白多肽绿色生物制造广东普通高校重点实验室,广东 深圳 518055
3.清华大学化工系生物化工研究所,工业生物催化教育部重点实验室,北京 100084
4.清华大学合成与系统生物学研究中心,北京 100084
5.华南理工大学生物科学与工程学院,广东 广州 510006
-
收稿日期:2024-11-11修回日期:2025-02-09出版日期:2025-05-25发布日期:2025-05-20 -
通讯作者:邢新会 -
作者简介:王媛媛(1997—),女,博士研究生,研究方向为合成生物学。E-mail:2573291738@qq.com。 -
基金资助:国家重点研发计划(2018YFA0901500);深圳市高等院校稳定支持计划(WDZC20231126172733002)
Research progress on bioproduction of recombinant proteins by Pichia pastoris utilizing methanol
WANG Yuanyuan1,2( ), ZHANG Chong3,4, HAN Shuangyan5, XING Xinhui1,2,3,4(
), ZHANG Chong3,4, HAN Shuangyan5, XING Xinhui1,2,3,4( )
)
- 1.Institute of Biopharmaceutical and Health Engineering, Tsinghua Shenzhen International Graduate School, Shenzhen 518055, Guangdong, China
2.Key Laboratory of Active Proteins and Peptides Green Biomanufacturing of Guangdong Higher Education Institutes, Tsinghua Shenzhen International Graduate School, Shenzhen 518055, Guangdong, China
3.Key Laboratory for Industrial Biocatalysis of Ministry of Education, Institute of Biochemical Engineering, Department of Chemical Engineering, Tsinghua University, Beijing 100084, China
4.Center for Synthetic and Systems Biology, Tsinghua University, Beijing 100084, China
5.School of Biology and Biological Engineering, South China University of Technology, Guangzhou 510006, Guangdong, China
-
Received:2024-11-11Revised:2025-02-09Online:2025-05-25Published:2025-05-20 -
Contact:XING Xinhui
摘要:
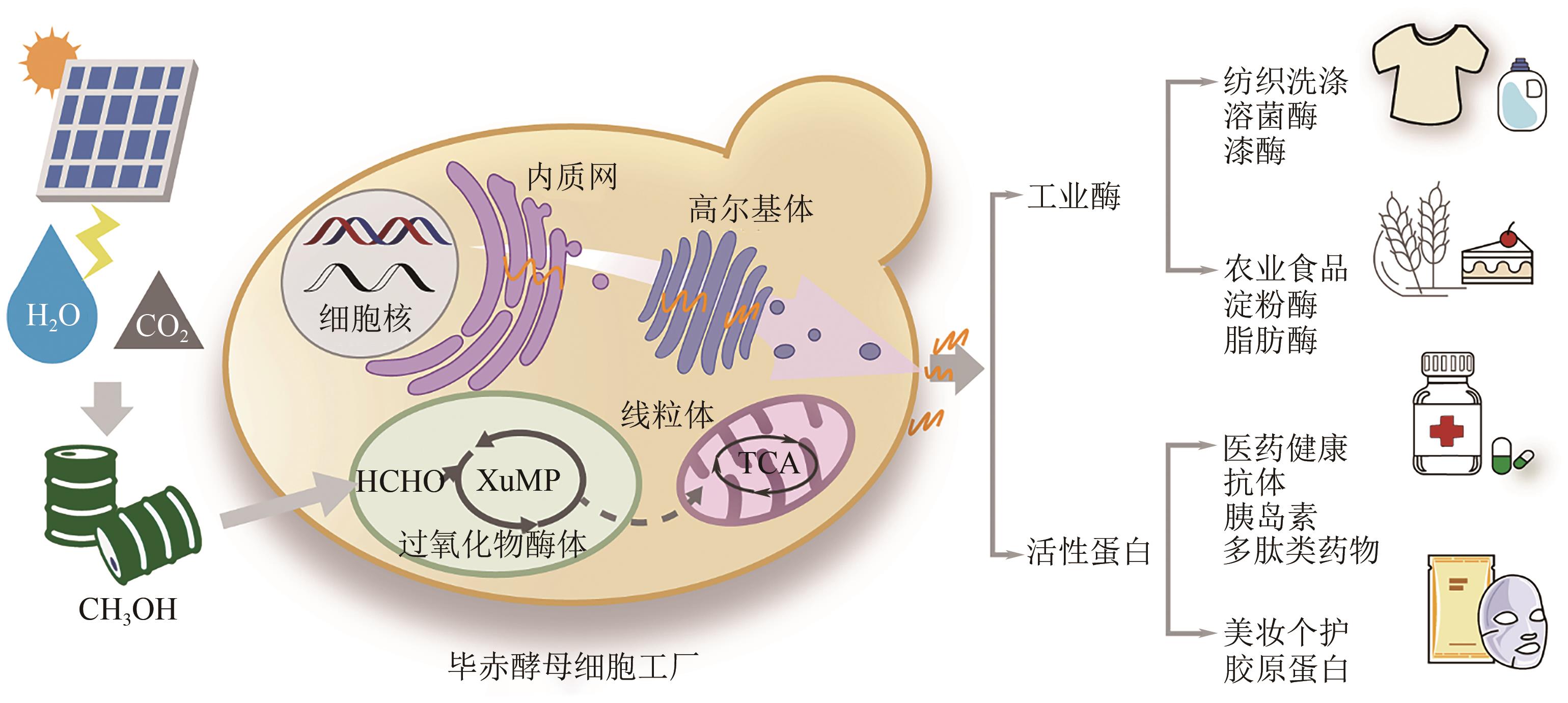
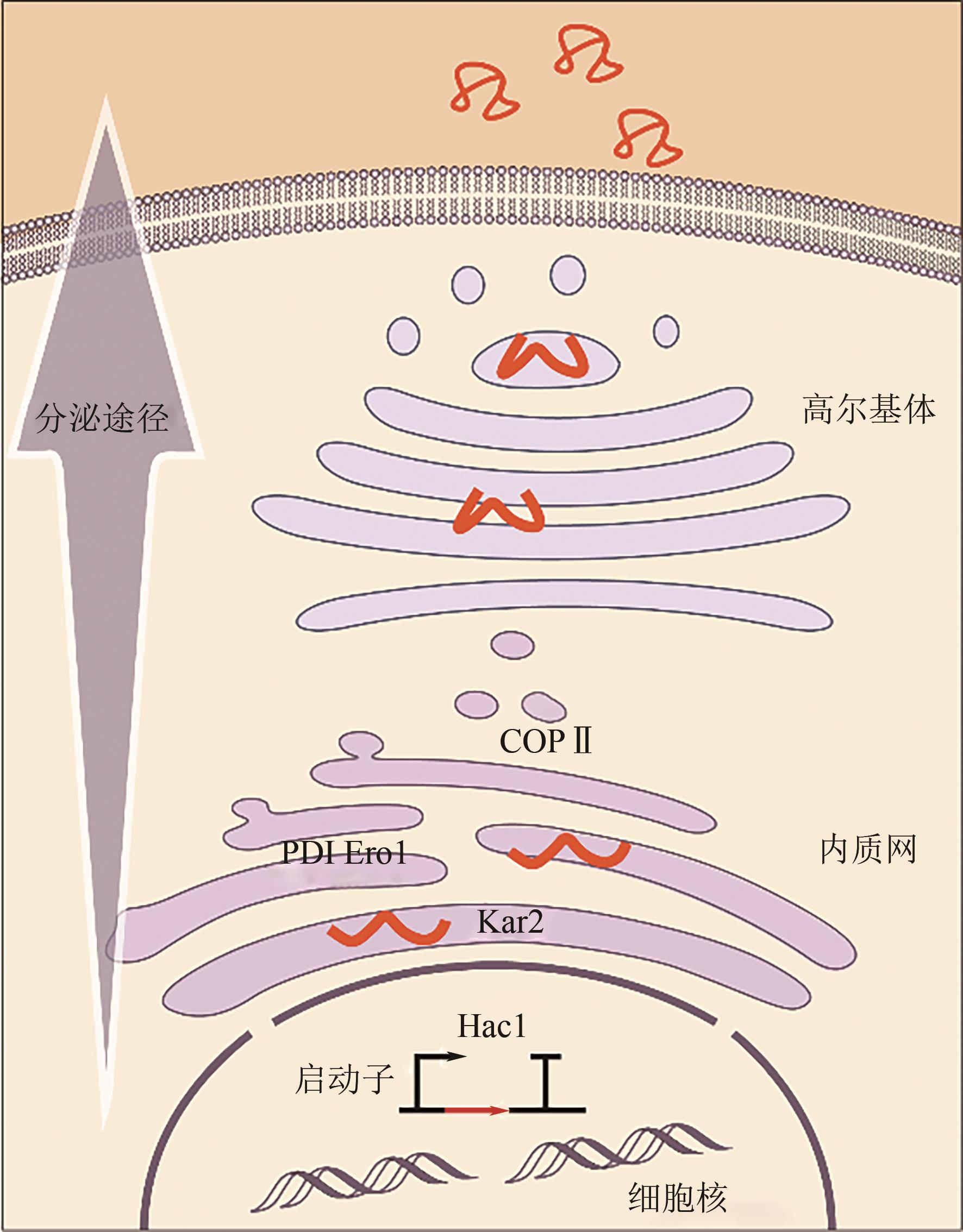
甲醇等一碳化合物作为生物制造的一种绿色可持续低碳原料,不存在与人争粮的问题,具有巨大的应用发展潜力。能够天然利用甲醇的毕赤酵母研究历史长,已被广泛应用于多种重组蛋白的生物制造中,成为甲醇利用性能优良的底盘细胞工厂。近年虽然对毕赤酵母的生理学、甲醇代谢网络、蛋白分泌表达途径及关键元件挖掘开展了大量的研究,积累了大量知识,但仍存在影响其高效应用的一系列关键问题,如甲醇及其代谢物对细胞毒性的复杂影响机制认知有限、细胞鲁棒性及甲醇利用相关的“生命暗物质”挖掘能力不足、甲醇高效转化成目标蛋白的工程化手段有限等。本文综述了国内外有关毕赤酵母生物转化甲醇合成重组蛋白的研究现状,重点总结讨论了毕赤酵母细胞工厂转化甲醇及蛋白表达优化策略,最后探讨了当前毕赤酵母生物转化甲醇生产重组蛋白过程中面临的挑战,并对其未来的生物技术创新和应用进行了展望。
中图分类号:
引用本文
王媛媛, 张翀, 韩双艳, 邢新会. 毕赤酵母利用甲醇生产重组蛋白技术的研究进展[J]. 化工进展, 2025, 44(5): 2441-2450.
WANG Yuanyuan, ZHANG Chong, HAN Shuangyan, XING Xinhui. Research progress on bioproduction of recombinant proteins by Pichia pastoris utilizing methanol[J]. Chemical Industry and Engineering Progress, 2025, 44(5): 2441-2450.
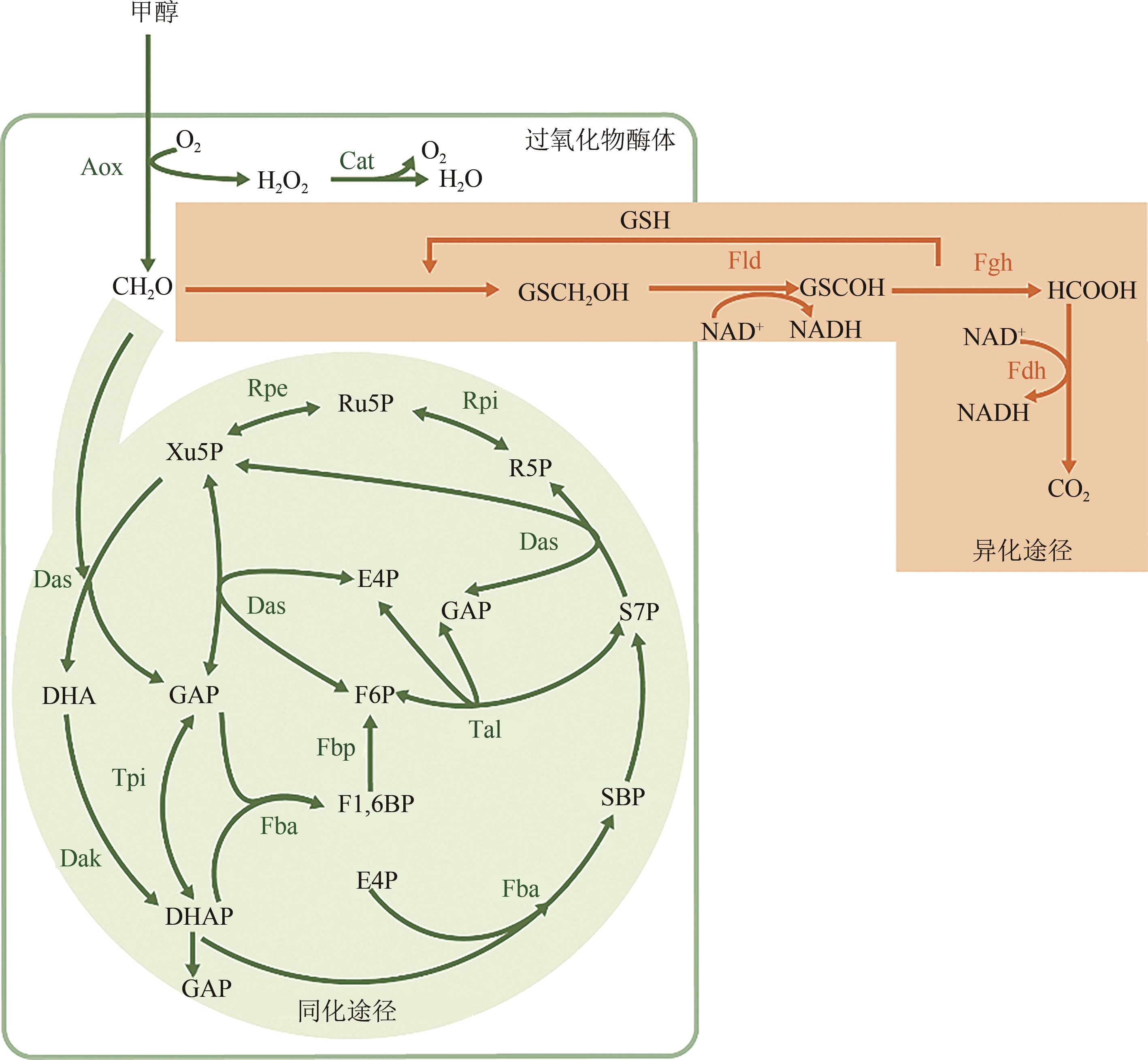
图2 毕赤酵母甲醇代谢途径Fld—甲醛脱氢酶;Fdh—甲酸脱氢酶;Fgh—S-羟甲基谷胱甘肽水解酶;Dak—二羟基丙酮激酶;Fbp—果糖二磷酸酶;Tpi—磷酸丙糖异构酶;Tal—转醛醇酶;Rpi—核糖-5-磷酸异构酶;Rpe—核酮糖磷酸3-差向异构酶;GAP—甘油醛-3-磷酸;E4P—赤藓糖-4-磷酸;SBP—景天庚酮糖-1,7-二磷酸;S7P—景天庚酮糖-7-磷酸;R5P—核糖-5-磷酸;Ru5P—核酮糖-5-磷酸;CH2O—甲醛;GSH—还原型谷胱甘肽;GSCH2OH—S-羟甲酰谷胱甘肽;GSCHO—S-羟甲基谷胱甘肽;HCOOH—甲酸
| 1 | 武云飞, 王莹莹, 朱家龙, 等. 绿色甲醇生产与应用协同发展实施路径探讨[J]. 煤炭加工与综合利用, 2024(9): 38-45. |
| WU Yunfei, WANG Yingying, ZHU Jialong, et al. Research on implementation pathway of coordinated development for green methanol production and application[J]. Coal Processing & Comprehensive Utilization, 2024(9): 38-45. | |
| 2 | GUIL-LÓPEZ R, MOTA N, LLORENTE J, et al. Methanol synthesis from CO2: A review of the latest developments in heterogeneous catalysis[J]. Materials, 2019, 12(23): 3902. |
| 3 | MALHOTRA R, MATHEW T, SURYA PRAKASH G K. George Andrew Olah:Across conventional lines[J]. Resonance, 2017, 22(12): 1111-1153. |
| 4 | FABARIUS Jonathan Thomas, WEGAT Vanessa, ROTH Arne, et al. Synthetic methylotrophy in yeasts: Towards a circular bioeconomy[J]. Trends in Biotechnology, 2021, 39(4): 348-358. |
| 5 | UNVER Yagmur, DAGCI Ibrahim. Komagataella phaffii (Pichia pastoris) as a powerful yeast expression system for biologics production[J]. Frontiers in Bioscience, 2024, 16(2): 19. |
| 6 | SCHWARZHANS Jan-Philipp, LUTTERMANN Tobias, GEIER Martina, et al. Towards systems metabolic engineering in Pichia pastoris [J]. Biotechnology Advances, 2017, 35(6): 681-710. |
| 7 | 蔡鹏, 吴晓燕, 解林峰, 等. 甲醇微生物转化最新进展[J]. 中国科学: 化学, 2024, 54(11): 2199-2218. |
| CAI Peng, WU Xiaoyan, XIE Linfeng, et al. Advances in microbial bioconversion of methanol[J]. Scientia Sinica Chimica, 2024, 54(11): 2199-2218. | |
| 8 | 刘康, 乔杨怡, 张尚杰, 等. 甲醇生物转化合成化学品的研究进展[J]. 生物工程学报, 2023, 39(6): 2430-2448. |
| LIU Kang, QIAO Yangyi, ZHANG Shangjie, et al. Advances in biotransformation of methanol into chemicals[J]. Chinese Journal of Biotechnology, 2023, 39(6): 2430-2448. | |
| 9 | HANSEN H, DIDION T, THIEMANN A, et al. Targeting sequences of the two major peroxisomal proteins in the methylotrophic yeast Hansenula polymorpha [J]. Molecular & General Genetics, 1992, 235(2/3): 269-278. |
| 10 | MÜLLER Jonas E N, LITSANOV Boris, Miriam BORTFELD-MILLER, et al. Proteomic analysis of the thermophilic methylotroph Bacillus methanolicus MGA3[J]. Proteomics, 2014, 14(6): 725-737. |
| 11 | 孙青, 刘德华, 陈振. 甲醇的生物利用与转化[J]. 中国生物工程杂志, 2020, 40(10): 65-75. |
| SUN Qing, LIU Dehua, CHEN Zhen. Research progress of methanol utilization and bioconversion[J]. China Biotechnology, 2020, 40(10): 65-75. | |
| 12 | WANG Yuanyuan, LI Jingwen, ZHAO Fengguang, et al. Methanol oxidase from Hansenula polymorpha shows activity in peroxisome-deficient Pichia pastoris [J]. Biochemical Engineering Journal, 2022, 180: 108369. |
| 13 | ROTH Timothy B, WOOLSTON Benjamin M, STEPHANOPOULOS Gregory, et al. Phage-assisted evolution of bacillus methanolicus methanol dehydrogenase 2[J]. ACS Synthetic Biology, 2019, 8(4): 796-806. |
| 14 | CAI Peng, WU Xiaoyan, DENG Jun, et al. Methanol biotransformation toward high-level production of fatty acid derivatives by engineering the industrial yeast Pichia pastoris [J]. Proceedings of the National Academy of Sciences of the United States of America, 2022, 119(29): e2201711119. |
| 15 | SHEN Yiwei, CAI Peng, GAO Linhui, et al. Engineering high production of fatty alcohols from methanol by constructing coordinated dual biosynthetic pathways[J]. Bioresource Technology, 2024, 412: 131396. |
| 16 | KRAINER Florian W, DIETZSCH Christian, HAJEK Tanja, et al. Recombinant protein expression in Pichia pastoris strains with an engineered methanol utilization pathway[J]. Microbial Cell Factories, 2012, 11: 22. |
| 17 | 韩双艳, 李静文, 王媛媛. 利用SpyTag/SpyCatcher体系提高毕赤酵母重组蛋白生产能力[J]. 华南理工大学学报(自然科学版), 2022, 50(1): 143-154. |
| HAN Shuangyan, LI Jingwen, WANG Yuanyuan. Production capacity improvement of recombinant proteins of Pichia pastoris with the SpyTag/SpyCatcher system[J]. Journal of South China University of Technology (Natural Science Edition), 2022, 50(1): 143-154. | |
| 18 | ZHAO Bingjie, LI Yu, ZHANG Yong, et al. Low-carbon and overproduction of cordycepin from methanol using engineered Pichia pastoris cell factory[J]. Bioresource Technology, 2024, 413: 131446. |
| 19 | WANG Yuanyuan, LI Ruisi, ZHAO Fengguang, et al. Metabolic engineering of Komagataella phaffii for the efficient utilization of methanol[J]. Microbial Cell Factories, 2024, 23(1): 198. |
| 20 | WOOLSTON Benjamin M, KING Jason R, REITER Michael, et al. Improving formaldehyde consumption drives methanol assimilation in engineered E. coli [J]. Nature Communications, 2018, 9(1): 2387. |
| 21 | YU Yifan, YANG Jiashuo, ZHAO Fengguang, et al. Comparative transcriptome and metabolome analyses reveal the methanol dissimilation pathway of Pichia pastoris [J]. BMC Genomics, 2022, 23(1): 366. |
| 22 | BOOJARI Mohammad Amin, GHALEDARI Fatemeh Rajabi, MOTAMEDIAN Ehsan, et al. Developing a metabolic model-based fed-batch feeding strategy for Pichia pastoris fermentation through fine-tuning of the methanol utilization pathway[J]. Microbial Biotechnology, 2023, 16(6): 1344-1359. |
| 23 | SOLTANI Maryam Khalifeh, ARJMAND Sareh, SIADAT Seyed Omid Ranaei, et al. Hansenula polymorpha methanol metabolism genes enhance recombinant protein production in Komagataella phaffi [J]. AMB Express, 2024, 14(1): 88. |
| 24 | 汤晓玲, 陈静祥, 柳志强, 等. 基于工业环境扰动的微生物适应性进化[J]. 生物工程学报, 2023, 39(3): 993-1008. |
| TANG Xiaoling, CHEN Jingxiang, LIU Zhiqiang, et al. Adaptive evolution of microorganisms based on industrial environmental perturbations[J]. Chinese Journal of Biotechnology, 2023, 39(3): 993-1008. | |
| 25 | MAVROMMATI Maria, DASKALAKI Alexandra, PAPANIKOLAOU Seraphim, et al. Adaptive laboratory evolution principles and applications in industrial biotechnology[J]. Biotechnology Advances, 2022, 54: 107795. |
| 26 | MENG Jiao, LIU Shufan, GAO Le, et al. Economical production of Pichia pastoris single cell protein from methanol at industrial pilot scale[J]. Microbial Cell Factories, 2023, 22(1): 198. |
| 27 | WANG Shuai, WANG Yuanyuan, YUAN Qingyan, et al. Development of high methanol-tolerance Pichia pastoris based on iterative adaptive laboratory evolution[J]. Green Chemistry, 2023, 25(21): 8845-8857. |
| 28 | 韩双艳, 王帅. 磷脂酰胆碱合成途径膜相关基因在增强毕赤酵母有机溶剂耐受性中的应用: CN118530863A[P]. 2024-08-23. |
| HAN Shuangyan, WANG Shuai, Application of membrane-associated genes in the phosphatidylcholine synthesis pathway in enhancing organic solvent tolerance in Pichia pastoris: CN118530863A[P]. 2024-08-23. | |
| 29 | 韩双艳, 王帅, 滕飞, 等. 一种耐受甲醇的高产α-亚麻酸的毕赤酵母工程菌及其构建方法和应用: CN118599690A[P]. 2024-09-06. |
| HAN Shuangyan, WANG Shuai, TENG Fei, et al. A methanol-tolerant Pichia pastoris engineered strain with high α-linolenic acid production, its construction method, and application: CN118599690A[P]. 2024-09-06. | |
| 30 | 韩双艳, 王帅, 张亚萍. 膜相关基因在提高毕赤酵母甲醇耐受性中的应用: CN118389309A[P]. 2024-07-26. |
| HAN Shuangyan, WANG Shuai, ZHANG Yaping. Application of membrane-associated genes in improving methanol tolerance in Pichia pastoris : CN118389309A[P]. 2024-07-26. | |
| 31 | 韩双艳, 王帅, 张亚萍. 内源膜相关基因在提高毕赤酵母甲醇耐受性中的应用: CN118374374A[P]. 2024-07-23. |
| HAN Shuangyan, WANG Shuai, ZHANG Yaping. Application of endogenous membrane-associated genes in. improving methanol tolerance in Pichia pastoris: CN118374374A[P]. 2024-07-23. | |
| 32 | 韩双艳, 王帅, 赵风光. 磷脂酰胆碱短合成途径膜相关基因在提高毕赤酵母甲醇耐受性中的应用: CN118374373A[P]. 2024-07-23. |
| HAN Shuangyan, WANG Shuai, ZHAO Fengguang. Application of membrane-associated genes in the short phosphatidylcholine synthesis pathway in improving methanol tolerance in Pichia pastoris : CN118374373A[P]. 2024-07-23. | |
| 33 | 韩双艳, 王帅, 赵风光. 心磷脂合成途径膜相关基因在提高毕赤酵母甲醇耐受性中的应用: CN118374375A[P]. 2024-07-23. |
| HAN Shuangyan, WANG Shuai, ZHAO Fengguang. Application of cardiolipin synthetic pathway membrane related gene in improving methanol tolerance of Pichia pastoris : CN118374375A[P]. 2024-07-23. | |
| 34 | 高乐, 吴信, 孟娇. 一种高甲醇耐受的毕赤酵母菌株的构建及其应用: CN115851473A[P]. 2023-03-28. |
| GAO Le, WU Xin, MENG Jiao. Construction and application of Pichia pastoris strain with high methanol tolerance: CN115851473A[P]. 2023-03-28. | |
| 35 | BARONE Giovanni Davide, Anita EMMERSTORFER-AUGUSTIN, BIUNDO Antonino, et al. Industrial production of proteins with Pichia pastoris—Komagataella phaffii [J]. Biomolecules, 2023, 13(3): 441. |
| 36 | 袁姚梦. 基于碱基编辑器的微生物细胞工厂合成进化[D]. 北京: 清华大学, 2024. |
| YUAN Yaomeng. Base editor-mediated synthetic evolution of microbial cell factories[D]. Beijing: Tsinghua University, 2024. | |
| 37 | ZHANG Siyu, LIN Ru, CUI Luyao, et al. Alter codon bias of the P. pastoris genome to overcome a bottleneck in codon optimization strategy development and improve protein expression[J]. Microbiological Research, 2024, 282: 127629. |
| 38 | 宋小平, 王雅洁, 蔡晶晶, 等. 基因拷贝数对重组毕赤酵母产谷氨酰胺转胺酶的影响[J]. 生物工程学报, 2020, 36(8): 1679-1688. |
| SONG Xiaoping, WANG Yajie, CAI Jingjing, et al. Impact of gene dosage on recombinant transglutaminase production of Pichia pastoris [J]. Chinese Journal of Biotechnology, 2020, 36(8): 1679-1688. | |
| 39 | PORTELA Rui M C, VOGL Thomas, KNIELY Claudia, et al. Synthetic core promoters as universal parts for fine-tuning expression in different yeast species[J]. ACS Synthetic Biology, 2017, 6(3): 471-484. |
| 40 | ZOU Chenwei, LU Lingfang, WANG Shengyan, et al. The α-mating factor secretion signals and endogenous signal peptides for recombinant protein secretion in Komagataella phaffii [J]. Biotechnology for Biofuels and Bioproducts, 2022, 15(1): 140. |
| 41 | KJELDSEN Thomas, ANDERSEN Asser Sloth, František HUBÁLEK, et al. Molecular engineering of insulin for recombinant expression in yeast[J]. Trends in Biotechnology, 2024, 42(4): 464-478. |
| 42 | BARRERO Juan J, CASLER Jason C, VALERO Francisco, et al. An improved secretion signal enhances the secretion of model proteins from Pichia pastoris [J]. Microbial Cell Factories, 2018, 17(1): 161. |
| 43 | RAEMAEKERS Romaan J M, DE MURO Laura, GATEHOUSE John A, et al. Functional phytohemagglutinin (PHA) and Galanthus nivalis agglutinin (GNA) expressed in Pichia pastoris [J]. European Journal of Biochemistry, 1999, 265(1): 394-403. |
| 44 | KARAOGLAN Mert, YILDIZ Hilal, INAN Mehmet. Screening of signal sequences for extracellular production of Aspergillus Niger xylanase in Pichia pastoris [J]. Biochemical Engineering Journal, 2014, 92: 16-21. |
| 45 | DUAN Guangdong, DING Lumei, WEI Dongsheng, et al. Screening endogenous signal peptides and protein folding factors to promote the secretory expression of heterologous proteins in Pichia pastoris [J]. Journal of Biotechnology, 2019, 306: 193-202. |
| 46 | ZAHRL Richard J, PEÑA David A, MATTANOVICH Diethard, et al. Systems biotechnology for protein production in Pichia pastoris [J]. FEMS Yeast Research, 2017, 17(7). |
| 47 | DAMASCENO Leonardo M, ANDERSON Kyle A, RITTER Gerd, et al. Cooverexpression of chaperones for enhanced secretion of a single-chain antibody fragment in Pichia pastoris [J]. Applied Microbiology and Biotechnology, 2007, 74(2): 381-389. |
| 48 | BAO Jichen, HUANG Mingtao, PETRANOVIC Dina, et al. Moderate expression of SEC16 increases protein secretion by Saccharomyces cerevisiae [J]. Applied and Environmental Microbiology, 2017, 83(14): e03400-16. |
| 49 | LI Jia, SHAO Qinan, XIANG Yulong, et al. High-activity recombinant human carboxypeptidase B expression in Pichia pastoris through rational protein engineering and enhancing secretion from the Golgi apparatus to the plasma membrane[J]. Biotechnology Journal, 2024, 19(5): 2400098. |
| 50 | Xueqin LYU, CUI Shixiu, CHEN Jie, et al. Cascaded de novo biosynthesis of lacto-proteins from CO2 by engineered Pichia pastoris [J]. Green Chemistry, 2023, 25(14): 5460-5469. |
| 51 | LI Feiran, CHEN Yu, QI Qi, et al. Improving recombinant protein production by yeast through genome-scale modeling using proteome constraints[J]. Nature Communications, 2022, 13(1): 2969. |
| 52 | WANG Tianmin, GUAN Changge, GUO Jiahui, et al. Pooled CRISPR interference screening enables genome-scale functional genomics study in bacteria with superior performance[J]. Nature Communications, 2018, 9(1): 2475. |
| 53 | YU Xinyu, LI Shuang, FENG Huibao, et al. CRISPRi-microfluidics screening enables genome-scale target identification for high-titer protein production and secretion[J]. Metabolic Engineering, 2023, 75: 192-204. |
| 54 | MORMINO Maurizio, LENITZ Ibai, SIEWERS Verena, et al. Identification of acetic acid sensitive strains through biosensor-based screening of a Saccharomyces cerevisiae CRISPRi library[J]. Microbial Cell Factories, 2022, 21(1): 214. |
| 55 | YANG Haiquan, QU Jinfeng, ZOU Wei, et al. An overview and future prospects of recombinant protein production in Bacillus subtilis [J]. Applied Microbiology and Biotechnology, 2021, 105(18): 6607-6626. |
| 56 | ZHU Zirong, DING Xuezhi, RANG Jie, et al. Application and research progress of ARTP mutagenesis in actinomycetes breeding[J]. Gene, 2024, 929: 148837. |
| 57 | CAI Peng, DUAN Xingpeng, WU Xiaoyan, et al. Recombination machinery engineering facilitates metabolic engineering of the industrial yeast Pichia pastoris [J]. Nucleic Acids Research, 2021, 49(13): 7791-7805. |
| 58 | GAO Jucan, XU Junhao, ZUO Yimeng, et al. Synthetic biology toolkit for marker-less integration of multigene pathways into Pichia pastoris via CRISPR/Cas9[J]. ACS Synthetic Biology, 2022, 11(2): 623-633. |
| 59 | GAO Jucan, YE Cuifang, CHENG Jintao, et al. Enhancing homologous recombination efficiency in Pichia pastoris for multiplex genome integration using short homology arms[J]. ACS Synthetic Biology, 2022, 11(2): 547-553. |
| [1] | 吴孟勤, 王佳瑶, 徐友强, 王钰. 化学-生物级联转化CO2合成单细胞蛋白研究进展[J]. 化工进展, 2025, 44(5): 2429-2440. |
| [2] | 高建刚, 姜亚鹏, 包宝青, 王书琦, 崔书明. 绿氢转化制绿色甲醇与绿氨[J]. 化工进展, 2025, 44(4): 1987-1997. |
| [3] | 朱国瑜, 葛棋, 付名利. 甲醇重整制氢催化剂耐久性评价和寿命预测方法[J]. 化工进展, 2025, 44(3): 1338-1346. |
| [4] | 左骥, 罗莉, 谢永锴, 陈文尧, 钱刚, 周兴贵, 段学志. 甲醇无氧脱氢制甲醛Cu催化剂的粒径效应[J]. 化工进展, 2025, 44(3): 1347-1354. |
| [5] | 韩英娜, 李丽, 张林子, 安金泽, 李文秀, 张弢. 离子液体萃取精馏分离甲醇-乙腈共沸物[J]. 化工进展, 2025, 44(2): 660-668. |
| [6] | 孙悦鹏, 孙延吉, 潘艳秋, 王成宇. 基于BO-LSTM的低温甲醇洗净化气CO2含量预测[J]. 化工进展, 2025, 44(2): 688-697. |
| [7] | 胡洋, 韩传军, 胡强, 李汶颖, 安全成, 苏洋, 武洪松, 袁果. 固体氧化物燃料电池用甲醇水蒸气重整反应器研究进展[J]. 化工进展, 2025, 44(1): 169-183. |
| [8] | 周渝, 唐甜, 熊子悠, 韦奇. 基于两级微通道分离工艺的甲醇制烯烃废水深度处理[J]. 化工进展, 2025, 44(1): 100-108. |
| [9] | 周渝, 夏太阳, 韦奇, 唐甜, 田磊. 微通道耦合反渗透膜串联处理甲醇制烯烃废水工艺优化[J]. 化工进展, 2024, 43(S1): 43-51. |
| [10] | 龙涛, 周锋, 张伟, 吴泓, 王建, 陈霖. CO-CO2体系制备氘代甲醇催化剂的合成与改性[J]. 化工进展, 2024, 43(8): 4411-4420. |
| [11] | 郭鹏, 李红伟, 李贵贤, 季东, 王东亮, 赵新红. 直接甲醇燃料电池阳极催化剂的失活机制及应对策略[J]. 化工进展, 2024, 43(7): 3812-3823. |
| [12] | 方峣, 刘雷, 高志华, 黄伟, 左志军. 光辅助直接甲醇燃料电池阳极催化剂的研究进展[J]. 化工进展, 2024, 43(5): 2611-2628. |
| [13] | 周运桃, 王洪星, 李新刚, 崔丽凤. CeO2载体在CO2加氢制甲醇中的应用和研究进展[J]. 化工进展, 2024, 43(5): 2723-2738. |
| [14] | 周秋明, 牛丛丛, 吕帅帅, 李红伟, 文富利, 徐润, 李明丰. 通过产物转化分离推动CO2加氢制甲醇过程的研究进展[J]. 化工进展, 2024, 43(5): 2776-2785. |
| [15] | 李海鹏, 吴桐, 王琪, 郜时旺, 王晓龙, 李旭, 高新华, 年佩, 魏逸彬. 透水NaA分子筛膜强化的CO2加氢高效制甲醇[J]. 化工进展, 2024, 43(5): 2834-2842. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||