化工进展 ›› 2021, Vol. 40 ›› Issue (3): 1215-1225.DOI: 10.16085/j.issn.1000-6613.2020-1933
D-氨基酸氧化酶的分子改造及应用研究进展
- 1.浙江大学化学工程与生物工程学院,浙江 杭州 310027
2.浙江大学杭州国际科创中心,浙江 杭州 311200
-
收稿日期:2020-09-22出版日期:2021-03-05发布日期:2021-03-17 -
通讯作者:杨立荣 -
作者简介:居述云(1988—),男,博士,研究方向为生物催化与转化。E-mail:jushuyun@zju.edu.cn 。 -
基金资助:国家重点研发计划(2019YFA09005000)
Advances in the molecular modification and application of D-amino acid oxidase
JU Shuyun1,2( ), WU Jianping1,2, YANG Lirong1,2(
), WU Jianping1,2, YANG Lirong1,2( )
)
- 1.College of Chemical and Biological Engineering, Zhejiang University, Hangzhou 310027, Zhejiang, China
2.Hangzhou Global Scientific and Technological Innovation Center, Zhejiang University, Hangzhou 311200, Zhejiang, China
-
Received:2020-09-22Online:2021-03-05Published:2021-03-17 -
Contact:YANG Lirong
摘要:
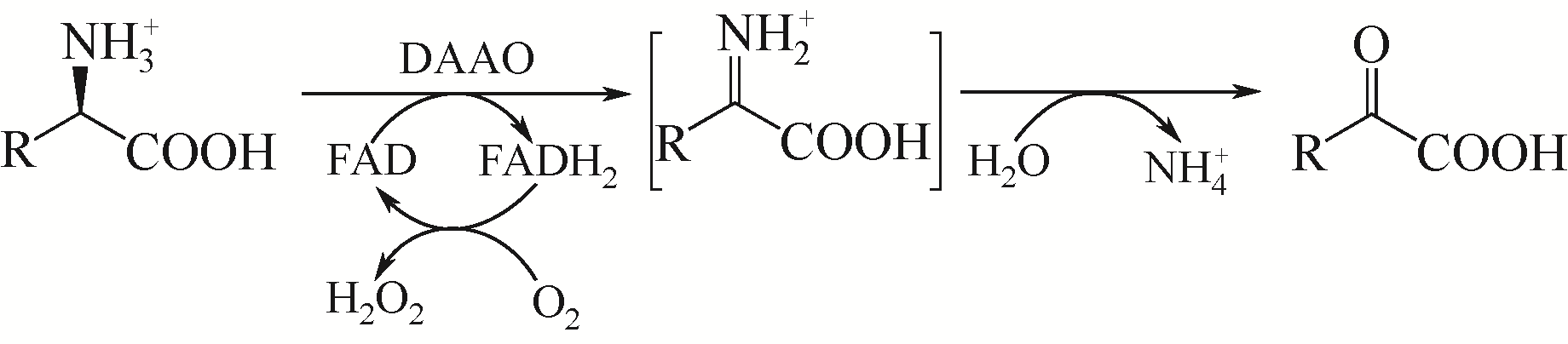
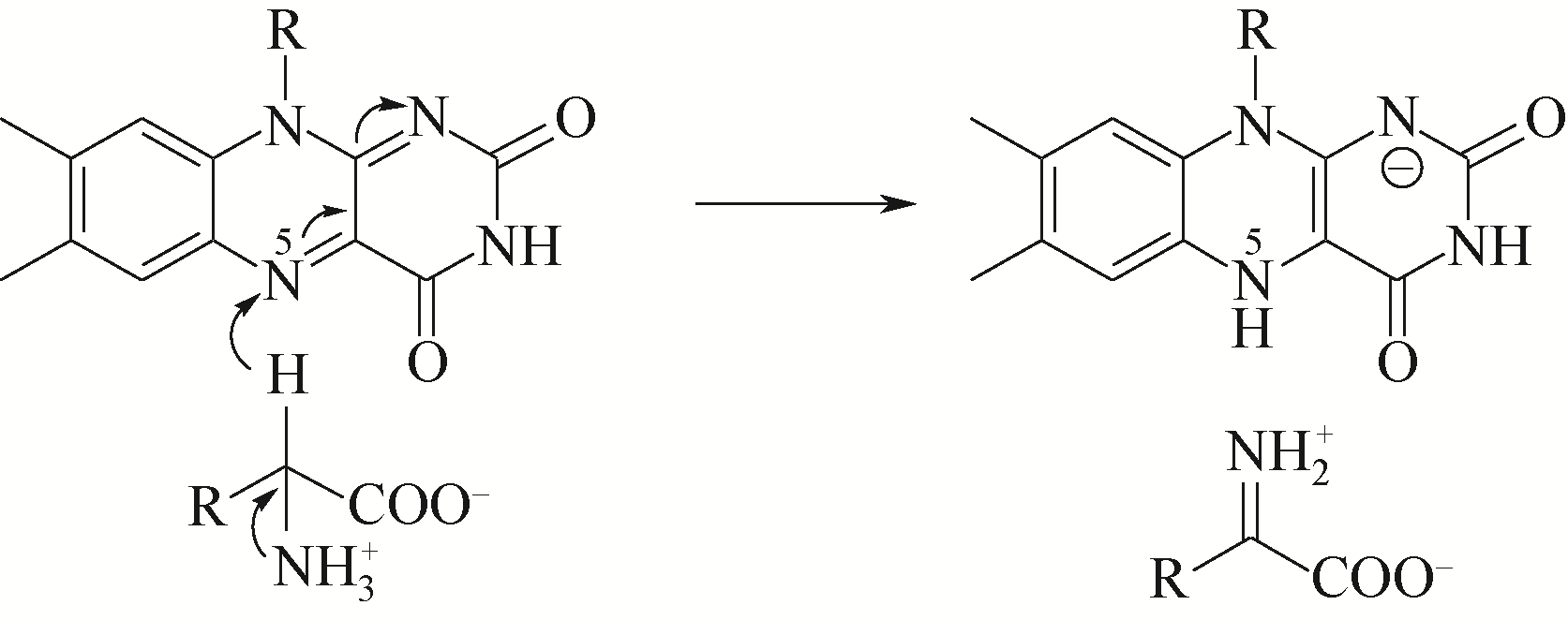
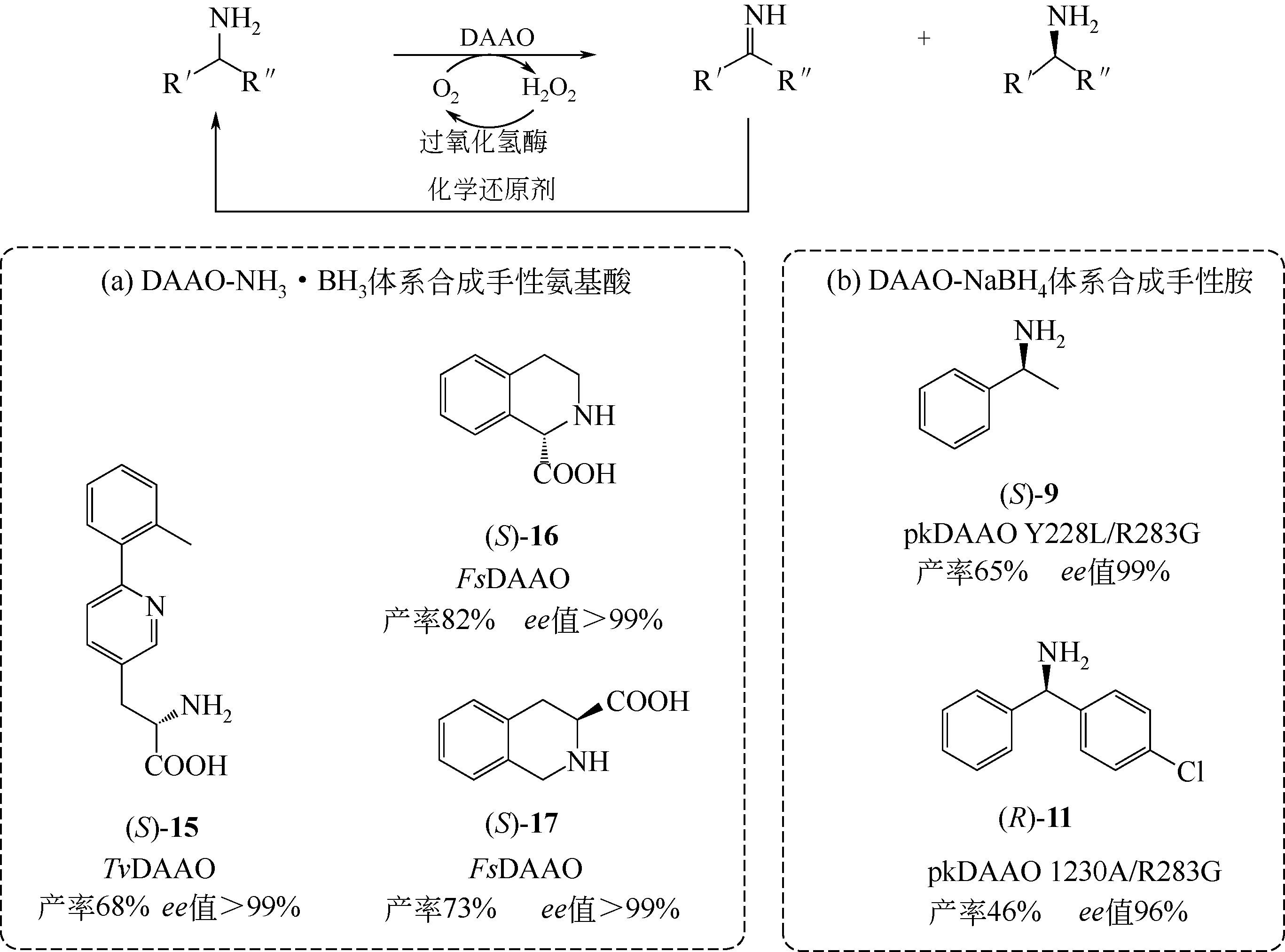
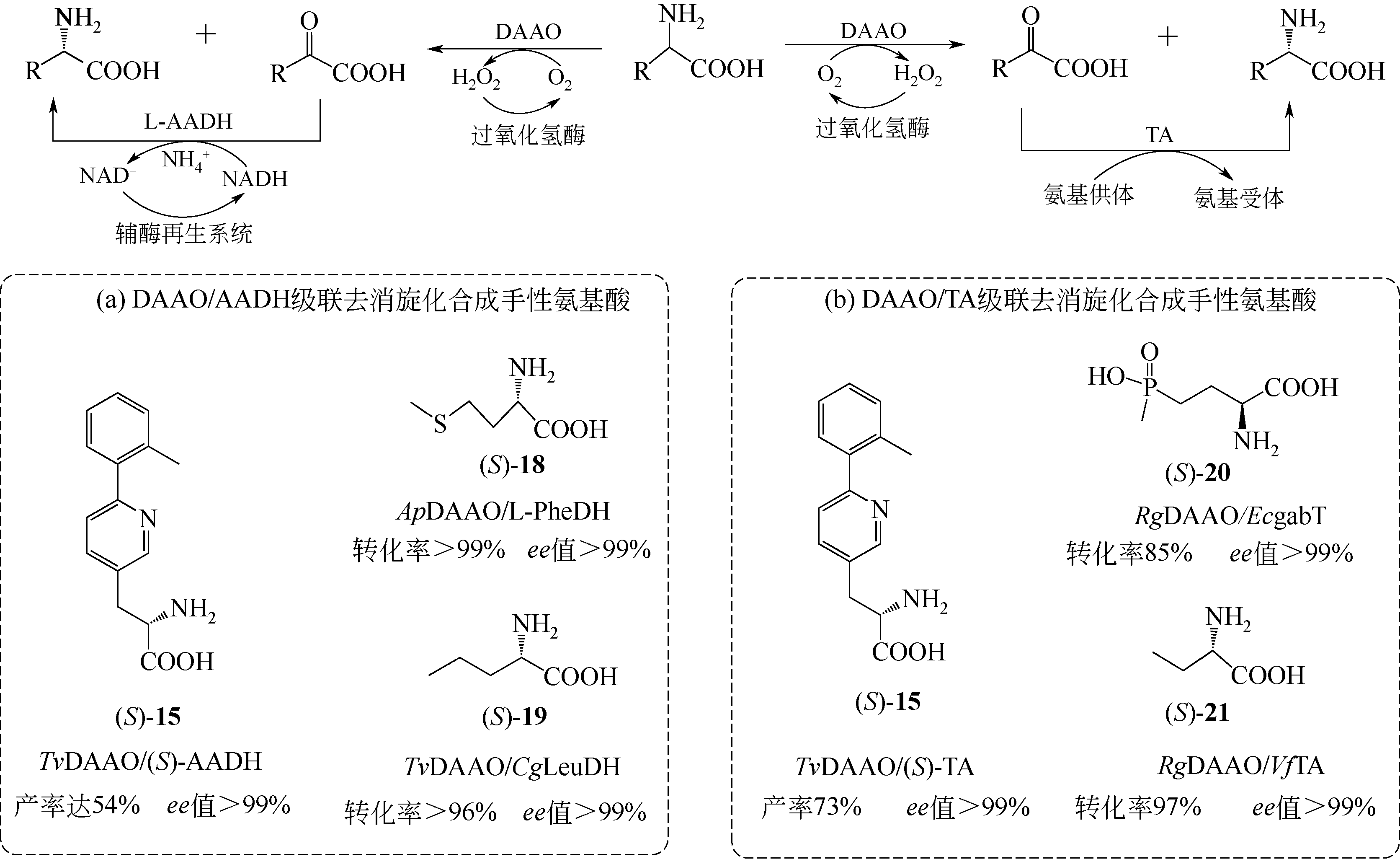
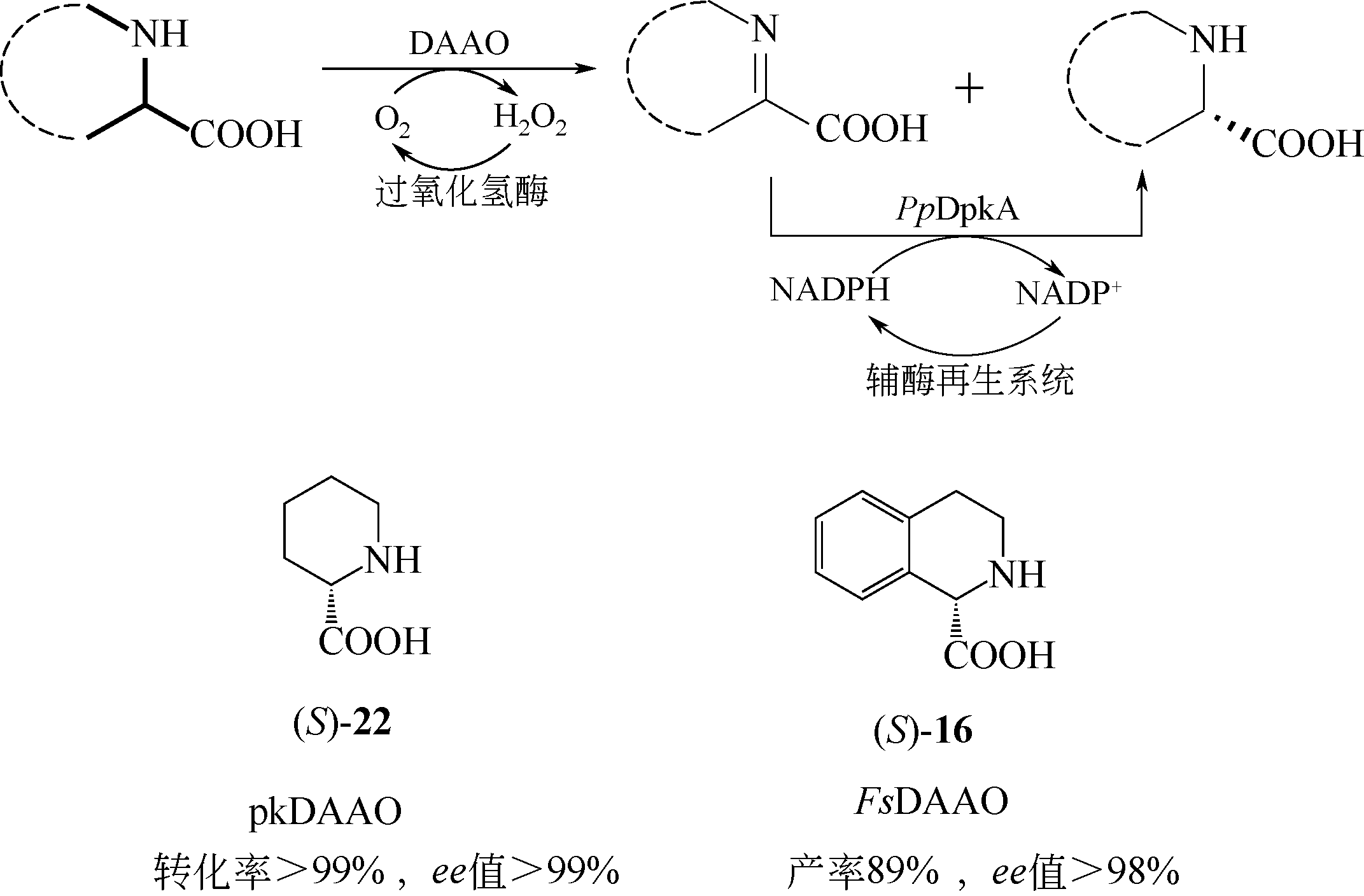
D-氨基酸氧化酶是一类含有黄素腺嘌呤二核苷酸的氧化还原酶,能够催化D-氨基酸氧化脱氢,生成相应的α-酮酸、过氧化氢和氨。该类酶在自然界中分布广泛,主要来源于真核生物和少数原核生物。作为一种经典的生物催化剂,D-氨基酸氧化酶具有反应条件温和、底物谱广泛、对映体选择性好等特点,在合成医药、农药和精细化学品等方面具有重要的应用价值。本文综述了D-氨基酸氧化酶的基本蛋白结构特征及其催化机制,重点介绍了D-氨基酸氧化酶底物特异性和热稳定性分子改造的策略和代表性成果以及该类酶在生物催化中的应用,例如制备7-氨基头孢烷酸、手性氨基酸、胺类化合物和α-酮酸。最后探讨了D-氨基酸氧化酶目前在生物催化应用过程中存在的问题。后续的研究可围绕新酶的挖掘与改造展开工作。基于对映体选择性和底物识别的分子机制,理性设计酶的催化性能,并以挖掘或改造获得的D-氨基酸氧化酶作为新酶元件,用于构建功能化学品生物合成新途径。
中图分类号:
引用本文
居述云, 吴坚平, 杨立荣. D-氨基酸氧化酶的分子改造及应用研究进展[J]. 化工进展, 2021, 40(3): 1215-1225.
JU Shuyun, WU Jianping, YANG Lirong. Advances in the molecular modification and application of D-amino acid oxidase[J]. Chemical Industry and Engineering Progress, 2021, 40(3): 1215-1225.
| 参数 | RgDAAO | pkDAAO | hDAAO |
|---|---|---|---|
| 单亚基氨基酸数量 | 368 | 347 | 347 |
| 与RgDAAO的序列一致性 | 100% | 28.73% | 27.73% |
| 亚基分子量 | 40000 | 39600 | 39600 |
| 亚基聚合状态 | 同源二聚体+2FAD | 同源二聚体+2FAD | 同源二聚体+2FAD |
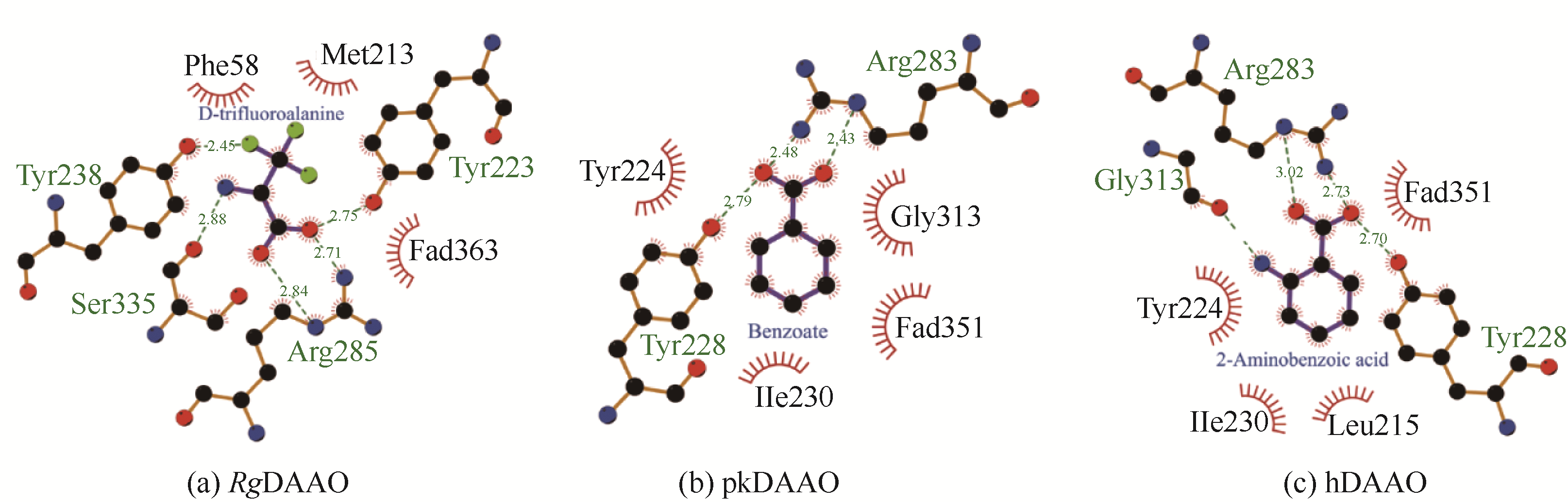
| 配体 | D-三氟丙氨酸 | 苯甲酸 | 2-氨基苯甲酸 |
| 与配体相互作用的关键氨基酸残基 | Y223,Y238,R285,S335, M213,F58 | Y224,Y228,R283, G313,I230 | Y224,Y228,R283,G313, I230,L215 |
| PDB登录号 | 1C0L | 1VE9 | 2E4A |
表1 RgDAAO、pkDAAO和hDAAO的基本性质比较
| 参数 | RgDAAO | pkDAAO | hDAAO |
|---|---|---|---|
| 单亚基氨基酸数量 | 368 | 347 | 347 |
| 与RgDAAO的序列一致性 | 100% | 28.73% | 27.73% |
| 亚基分子量 | 40000 | 39600 | 39600 |
| 亚基聚合状态 | 同源二聚体+2FAD | 同源二聚体+2FAD | 同源二聚体+2FAD |
| 配体 | D-三氟丙氨酸 | 苯甲酸 | 2-氨基苯甲酸 |
| 与配体相互作用的关键氨基酸残基 | Y223,Y238,R285,S335, M213,F58 | Y224,Y228,R283, G313,I230 | Y224,Y228,R283,G313, I230,L215 |
| PDB登录号 | 1C0L | 1VE9 | 2E4A |
| 原始酶 | 改造策略 | 突变体 | 底物 | kcat /min-1 | Km /mmol·L-1 | (kcat/Km) /mL·mol-1·min-1 | 参考 文献 |
|---|---|---|---|---|---|---|---|
| RgDAAO | 改变底物结合口袋处氨基酸残基的极性和带电性质,稳定底物结合构象 | M213R |  | 235(29) | 2(18) | 118(1.6) | [ |
 | 975(60) | 33(77.3) | 29.5(0.8) | ||||
 | 630(5000) | 17.8(0.8) | 35.4(6100) | ||||
| 减小底物结合口袋处氨基酸残基的空间位阻,扩大底物结合口袋 | M213G |  | 870(125) | 0.03(0.04) | 29000(3100) | [ | |
 | 31(6) | 0.03(0.01) | 1035(600) | ||||
 | 92(53) | 0.05(0.33) | 1840(160) | ||||
 | 950(180) | 1.6(2.1) | 600(90) | [ | |||
| 定向进化 | L118H |  | 3620(3900) | 0.4(0.9) | 8415(4330) | [ | |
 | 31(40) | 12.8(33.1) | 2.4(1.2) | ||||
T60A/Q144R /K152E |  | 53(40) | 7.9(33.1) | 6.7(1.2) | |||
| TvDAAO | 基于同源建模,改变底物结合口袋处氨基酸残基的极性,稳定底物结合构象 | F54Y |  | 2200(370) | 4.8(1.6) | 470(230) | [ |
| pkDAAO | 突变与底物(配体)羧基相互作用的精氨酸残基,重塑底物结合口袋 | Y228L/R283G |  | 444.0(-) | 7.0(-) | 63.4(-) | [ |
 | 46.8(-) | 1.5(-) | 31.2(-) | [ | |||
| 减小底物结合口袋处氨基酸残基的空间位阻,扩大底物结合口袋 | Y228L/R283G /F242I |  | 180.0(-) | 3.6(-) | 50.0(-) | [ | |
| I230A/R283G |  | 360.6(-) | 2.94(-) | 122.7(-) | [ |
表2 DAAO底物特异性改造的代表性成果
| 原始酶 | 改造策略 | 突变体 | 底物 | kcat /min-1 | Km /mmol·L-1 | (kcat/Km) /mL·mol-1·min-1 | 参考 文献 |
|---|---|---|---|---|---|---|---|
| RgDAAO | 改变底物结合口袋处氨基酸残基的极性和带电性质,稳定底物结合构象 | M213R |  | 235(29) | 2(18) | 118(1.6) | [ |
 | 975(60) | 33(77.3) | 29.5(0.8) | ||||
 | 630(5000) | 17.8(0.8) | 35.4(6100) | ||||
| 减小底物结合口袋处氨基酸残基的空间位阻,扩大底物结合口袋 | M213G |  | 870(125) | 0.03(0.04) | 29000(3100) | [ | |
 | 31(6) | 0.03(0.01) | 1035(600) | ||||
 | 92(53) | 0.05(0.33) | 1840(160) | ||||
 | 950(180) | 1.6(2.1) | 600(90) | [ | |||
| 定向进化 | L118H |  | 3620(3900) | 0.4(0.9) | 8415(4330) | [ | |
 | 31(40) | 12.8(33.1) | 2.4(1.2) | ||||
T60A/Q144R /K152E |  | 53(40) | 7.9(33.1) | 6.7(1.2) | |||
| TvDAAO | 基于同源建模,改变底物结合口袋处氨基酸残基的极性,稳定底物结合构象 | F54Y |  | 2200(370) | 4.8(1.6) | 470(230) | [ |
| pkDAAO | 突变与底物(配体)羧基相互作用的精氨酸残基,重塑底物结合口袋 | Y228L/R283G |  | 444.0(-) | 7.0(-) | 63.4(-) | [ |
 | 46.8(-) | 1.5(-) | 31.2(-) | [ | |||
| 减小底物结合口袋处氨基酸残基的空间位阻,扩大底物结合口袋 | Y228L/R283G /F242I |  | 180.0(-) | 3.6(-) | 50.0(-) | [ | |
| I230A/R283G |  | 360.6(-) | 2.94(-) | 122.7(-) | [ |
| 1 | 郭姣洁, 薛永常, 徐书景, 等. D-氨基酸氧化酶研究进展[J]. 中国生物工程杂志, 2010, 30(11): 106-111. |
| GUO Jiaojie, XUE Yongchang, XU Shujing, et al. D-amino acid oxidase update and review[J]. China Biotechnology, 2010, 30(11): 106-111. | |
| 2 | KREBS H A. Metabolism of amino-acids: deamination of amino-acids[J]. Biochemical Journal, 1935, 29(7): 1620-1644. |
| 3 | SIMONETTA M P, VANONI M A, CASALIN P. Purification and properties of D-amino acid oxidase, an inducible flavoenzyme from Rhodotorula gracilis[J]. Biochimica et Biophysica Acta: Protein Structure and Molecular Enzymology, 1987, 914(2): 136-142. |
| 4 | GONZÁLEZ F J, MONTES J, MARTIN F, et al. Molecular cloning of TvDAO1, a gene encoding a D-amino acid oxidase from Trigonopsis variabilis and its expression in Saccharomyces cerevisiae and Kluyveromyces lactis[J]. Yeast, 1997, 13(15): 1399-1408. |
| 5 | TAKAHASHI S, ABE K, KERA Y. Bacterial D-amino acid oxidases: recent findings and future perspectives[J]. Bioengineered, 2015, 6(4): 237-241. |
| 6 | POLLEGIONI L, PIUBELLI L, SACCHI S, et al. Physiological functions of D-amino acid oxidases: from yeast to humans[J]. Cellular and Molecular Life Sciences, 2007, 64(11): 1373-1394. |
| 7 | POLLEGIONI L, MOLLA G. New biotech applications from evolved D-amino acid oxidases[J]. Trends in Biotechnology, 2011, 29(6): 276-283. |
| 8 | BATALLA P, MARTIN A, LOPEZ M A, et al. Enzyme-based microfluidic chip coupled to graphene electrodes for the detection of D-amino acid enantiomer-biomarkers[J]. Analytical Chemistry, 2015, 87(10): 5074-5078. |
| 9 | UMHAU S, POLLEGIONI L, MOLLA G, et al. The X-ray structure of D-amino acid oxidase at very high resolution identifies the chemical mechanism of flavin-dependent substrate dehydrogenation[J]. Proceedings of the National Academy of Sciences, 2000, 97(23): 12463-12468. |
| 10 | POLLEGIONI L, DIEDERICHS K, MOLLA G, et al. Yeast D-amino acid oxidase: structural basis of its catalytic properties[J]. Journal of Molecular Biology, 2002, 324(3): 535-546. |
| 11 | MATTEVI A, CURTI B. Crystal structure of D-amino acid oxidase: a case of active site mirror-image convergent evolution with flavocytochrome b2[J]. Proceedings of the National Academy of Sciences, 1996, 93(15): 7496-7501. |
| 12 | MIZUTANI H, MIYAHARA I, HIROTSU K, et al. Three-dimensional structure of porcine kidney D-amino acid oxidase at 3.0Å resolution[J]. Journal of Biochemistry, 1996, 120(1): 14-17. |
| 13 | KAWAZOE T, TSUGE H, IMAGAWA T, et al. Structural basis of D-DOPA oxidation by D-amino acid oxidase: alternative pathway for dopamine biosynthesis[J]. Biochemical and Biophysical Research Communications, 2007, 355(2): 385-391. |
| 14 | POLLEGIONI L, MOLLA G, SACCHI S, et al. Properties and applications of microbial D-amino acid oxidases: current state and perspectives[J]. Applied Microbiology and Biotechnology, 2008, 78(1): 1-16. |
| 15 | 赵冉冉, 冯利伟, 张金秀, 等. D-氨基酸氧化酶结构的研究进展[J]. 生物技术通报, 2013(12): 27-35. |
| ZHAO Ranran, FENG Liwei, ZHANG Jinxiu, et al. Research progress on the crystal structure of D-amino acid oxidase[J]. Biotechnology Bulletin, 2013(12): 27-35. | |
| 16 | WALSH C T, SCHONBRUNN A, ABELES R H. Studies on the mechanism of action of D-amino acid oxidase. evidence for removal of substrate-hydrogen as a proton[J]. Journal of Biological Chemistry, 1971, 246(22): 6855-6866. |
| 17 | HERSH L B, JORNS M S. Use of 5-deazaFAD to study hydrogen transfer in the D-amino acid oxidase reaction[J]. Journal of Biological Chemistry, 1975, 250(22): 8728-8734. |
| 18 | DIJKMAN W P, DE GONZALO G, MATTEVI A, et al. Flavoprotein oxidases: classification and applications[J]. Applied Microbiology and Biotechnology, 2013, 97(12): 5177-88. |
| 19 | HARRIS C M, MOLLA G, PILONE M S, et al. Studies on the reaction mechanism of Rhodotorula gracilis D-amino-acid oxidase. role of the highly conserved Tyr-223 on substrate binding and catalysis[J]. Journal of Biological Chemistry, 1999, 274(274): 36233-36240. |
| 20 | SACCHI S, LORENZI S, MOLLA G, et al. Engineering the substrate specificity of D-amino acid oxidase[J]. Journal of Biological Chemistry, 2002, 277(30): 27510-27516. |
| 21 | YASUKAWA K, NAKANO S, ASANO Y. Tailoring D-amino acid oxidase from the pig kidney to R-stereoselective amine oxidase and its use in the deracemization of alpha-methylbenzylamine[J]. Angewandte Chemie: International Edition, 2014, 53(17): 4428-4431. |
| 22 | CALIGIURI A, D'ARRIGO P, ROSINI E, et al. Enzymatic conversion of unnatural amino acids by yeast D-amino acid oxidase[J]. Advanced Synthesis & Catalysis, 2006, 348(15): 2183-2190. |
| 23 | CALIGIURI A, D’ARRIGO P, ROSINI E, et al. Activity of yeast D-amino acid oxidase on aromatic unnatural amino acids[J]. Journal of Molecular Catalysis B: Enzymatic, 2008, 50(4): 93-98. |
| 24 | TRIMMER E E, WANNINAYAKE U S, FITZPATRICK P F. Mechanistic studies of an amine oxidase derived from D-amino acid oxidase[J]. Biochemistry, 2017, 56(14): 2024-2030. |
| 25 | NAKANO S, YASUKAWA K, TOKIWA T, et al. Origin of stereoselectivity and substrate/ligand recognition in an FAD-dependent R-selective amine oxidase[J]. Journal of Physical Chemistry B, 2016, 120: 10736-10743 |
| 26 | YASUKAWA K, MOTOJIMA F, ONO A, et al. Expansion of the substrate specificity of porcine kidney D-amino acid oxidase for S-stereoselective oxidation of 4-Cl-benzhydrylamine[J]. ChemCatChem, 2018, 10: 1-7. |
| 27 | WONG K S, FONG W P, TSANG P W. A single Phe54Tyr substitution improves the catalytic activity and thermostability of Trigonopsis variabilis D-amino acid oxidase[J]. New Biotechnology, 2010, 27(1): 78-84. |
| 28 | SACCHI S, ROSINI E, MOLLA G, et al. Modulating D-amino acid oxidase substrate specificity: production of an enzyme for analytical determination of all D-amino acids by directed evolution[J]. Protein Engineering, Design & Selection, 2004, 17(6): 517-525. |
| 29 | BAKKE M, SETOYAMA C, MIURA R, et al. Thermostabilization of porcine kidney D-amino acid oxidase by a single amino acid substitution[J]. Biotechnology and Bioengineering, 2006, 93(5): 1023-1027. |
| 30 | VOLPATO G, RODRIGUES R C, FERNANDEZ-LAFUENTE R. Use of enzymes in the production of semi-synthetic penicillins and cephalosporins: drawbacks and perspectives[J]. Current Medical Chemistry, 2010, 17(32): 3855-3873. |
| 31 | CONTI G, POLLEGIONI L, ROSINI E. One-pot conversion of cephalosporin C by using an optimized two-enzyme process[J]. Catalysis Science & Technology, 2015, 5(3): 1854-1863. |
| 32 | TAN Q, QIU J, LUO X, et al. Progress in one-pot bioconversion of cephalosporin C to 7-aminocephalosporanic acid[J]. Current Pharmaceutical Biotechnology, 2018, 19: 30-42. |
| 33 | HU L, MAGESH S, CHEN L, et al. Discovery of a small-molecule inhibitor and cellular probe of Keap1-Nrf2 protein-protein interaction[J]. Bioorganic & Medicinal Chemistry Letters, 2013, 23(10): 3039-3043. |
| 34 | KOTHA S, DEODHAR D, KHEDKAR P. Diversity-oriented synthesis of medicinally important 1,2,3,4-tetrahydroisoquinoline-3-carboxylic acid (Tic) derivatives and higher analogs[J]. Organic & Biomolecular Chemistry,2014, 12: 9054-9091. |
| 35 | PARK E, KIM M, J-S SHIN. One-pot conversion of L-threonine into L-homoalanine: biocatalytic production of an unnatural amino acid from a natural one[J]. Advanced Synthesis & Catalysis, 2010, 352(18): 3391-3398. |
| 36 | ENRIGHT A, F-R ALEXANDRE, ROFF G, et al. Stereoinversion of β- and γ-substituted α-amino acids using a chemo-enzymatic oxidation–reduction procedure[J]. Chemical Communications, 2003, 20: 2636-2637. |
| 37 | BEARD T M, TURNER N J. Deracemisation and stereoinversion of α-amino acids using D-amino acid oxidase and hydride reducing agents[J]. Chemical Communications, 2002, 3: 246-247. |
| 38 | ALEXANDRE F R, PANTALEONE D P, TAYLOR P P, et al. Amine-boranes: effective reducing agents for the deracemisation of DL-amino acids using L-amino acid oxidase from Proteus myxofaciens[J]. Tetrahedron Letters, 2002, 43(4): 707-710. |
| 39 | CHEN Y, GOLDBERG S L, HANSON R L, et al. Enzymatic preparation of an (S)-amino acid from a racemic amino acid[J]. Organic Process Research & Development, 2011, 15(1): 241-248. |
| 40 | JU S Y, QIAN M X, XU G, et al. Chemoenzymatic approach to (S)-1,2,3,4-tetrahydroisoquinoline carboxylic acids employing D-amino acid oxidase[J]. Advanced Synthesis & Catalysis, 2019, 361(13): 3191-3199. |
| 41 | FINDRIK Z, VASIC-RACKI D. Biotransformation of D-methionine into L-methionine in the cascade of four enzymes[J]. Biotechnology and Bioengineering, 2007, 98(5): 956-967. |
| 42 | QI Y, YANG T, ZHOU J, et al. Development of a multi-enzymatic desymmetrization and its application for the biosynthesis of L-norvaline from DL-norvaline[J]. Process Biochemistry, 2017, 55: 104-109. |
| 43 | GREEN B M, GRADLEY M L. Methods for making L-glufosinate: US 20170253897 A1[P]. 2017-09-07. |
| 44 | Y-M SEO, MATHEW S, H-S BEA, et al. Deracemization of unnatural amino acid: homoalanine using D-amino acid oxidase and ω-transaminase[J]. Organic & Biomolecular Chemistry, 2012, 10(12): 2482-2485. |
| 45 | LIU W, TANG D, SHI R, et al. Efficient production of S‐adenosyl‐L‐methionine from DL‐methionine in metabolic engineered Saccharomyces cerevisiae[J]. Biotechnology and Bioengineering, 2019, 116(12): 3312-3323. |
| 46 | YASUDA M, UEDA M, MURAMATSU H, et al. Enzymatic synthesis of cyclic amino acids by N-methyl-L-amino acid dehydrogenase from Pseudomonas putida[J]. Tetrahedron: Asymmetry, 2006, 17(12): 1775-1779. |
| 47 | JU S Y, QIAN M X, LI J, et al. A biocatalytic redox cascade approach for one-pot deracemization of carboxyl-substituted tetrahydroisoquinolines by stereoinversion[J]. Green Chemistry, 2019, 21(20): 5579-5585. |
| 48 | ORREGO A H, LÓPEZ-GALLEGO F, ESPAILLAT A, et al. One-step synthesis of alpha-keto acids from racemic amino acids by a versatile immobilized multienzyme cell-free system[J]. ChemCatChem, 2018, 10: 3002-3011. |
| 49 | MOLLA G, MELIS R, POLLEGIONI L. Breaking the mirror: L-amino acid deaminase, a novel stereoselective biocatalyst[J]. Biotechnology Advances, 2017, 35(6): 657-668. |
| 50 | HOSSAIN G S, LI J, SHIN H D, et al. L-amino acid oxidases from microbial sources: types, properties, functions, and applications[J]. Applied Microbiology and Biotechnology, 2014, 98(4): 1507-1515. |
| 51 | POLLEGIONI L, MOTTA P, MOLLA G. L-amino acid oxidase as biocatalyst: a dream too far?[J]. Applied Microbiology and Biotechnology, 2013, 97(21): 9323-9341. |
| 52 | SONG W, XU X, GAO C, et al. Open gate of corynebacterium glutamicum threonine deaminase for efficient synthesis of bulky α-keto acids[J]. ACS Catalysis, 2020, 10: 9994-10004. |
| [1] | 李华华, 李逸航, 金北辰, 李隆昕, 成少安. 厌氧氨氧化-生物电化学耦合废水处理系统的研究进展[J]. 化工进展, 2023, 42(5): 2678-2690. |
| [2] | 刘艳辉, 周明芳, 马铭, 王凯, 谭天伟. 可再生能源驱动的生物催化固定CO2的研究进展[J]. 化工进展, 2023, 42(1): 1-15. |
| [3] | 孟令玎, 毛梦雷, 廖奇勇, 孟子晖, 刘文芳. 碳酸酐酶和甲酸脱氢酶的稳定性研究进展[J]. 化工进展, 2022, 41(S1): 436-447. |
| [4] | 高博, 冯旭东, 李春. 可视化高通量检测天冬氨酸转氨甲酰酶活性的方法[J]. 化工进展, 2022, 41(4): 2054-2059. |
| [5] | 张彦, 汪伟, 谢锐, 巨晓洁, 刘壮, 褚良银. 负载酶@ZIF-8复合物的聚合物微颗粒可控制备[J]. 化工进展, 2022, 41(4): 2022-2028. |
| [6] | 李庆远, 王超, 许世佩, 张雪琴, 邱明建, 刘梦瑶, 丛梦晓. PBS前体1,4-丁二醇合成的反应工艺和催化剂研究进展[J]. 化工进展, 2022, 41(11): 5771-5782. |
| [7] | 鲁泽平, 裴新华, 薛誉, 张晓光, 胡燚. 甜菜碱类离子液体化学修饰猪胰脂肪酶提升其酶学性能[J]. 化工进展, 2022, 41(11): 6045-6052. |
| [8] | 李青, 刘武军, 郭潇佳, 王倩, 赵宗保. 手性NAD类似物合成及其辅酶应用[J]. 化工进展, 2021, 40(9): 5214-5221. |
| [9] | 张晓健, 刘倩, 柳志强, 郑裕国. 立体选择性羰基还原酶及其在手性醇合成中的应用[J]. 化工进展, 2021, 40(3): 1142-1160. |
| [10] | 赵婧, 王盼, 刘彦楠, 傅荣湛, 段志广, 范代娣. 人参皂苷的定向生物转化研究进展[J]. 化工进展, 2021, 40(3): 1238-1247. |
| [11] | 林朱凡, 成少安, 毛政中, 顾若男, 羊家威. 生物电化学脱氮系统构建和影响因素的最新研究进展[J]. 化工进展, 2020, 39(9): 3766-3776. |
| [12] | 王琛, 赵猛, 丁明珠, 王颖, 姚明东, 肖文海. 生物支架系统在合成生物学中的应用[J]. 化工进展, 2020, 39(11): 4557-4567. |
| [13] | 朱诚,许国超,戴威,周婕妤,倪晔. 醇脱氢酶KpADH的127位点对催化活性和对映选择性的影响[J]. 化工进展, 2019, 38(12): 5504-5511. |
| [14] | 姜恬, 冯旭东, 李岩, 李春. 底物特异性的生物催化与酶设计改造[J]. 化工进展, 2019, 38(01): 606-614. |
| [15] | 于波, 刘超, 刘金东, 丁万昱, 柴卫平. 介孔磷酸锆固体酸催化剂的制备及其催化性能[J]. 化工进展, 2018, 37(06): 2236-2241. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||