化工进展 ›› 2021, Vol. 40 ›› Issue (3): 1202-1214.DOI: 10.16085/j.issn.1000-6613.2020-2101
微生物合成植物天然产物的细胞工厂设计与构建
- 清华大学化学工程系,北京 100084
-
收稿日期:2020-10-20出版日期:2021-03-05发布日期:2021-03-17 -
通讯作者:李春 -
作者简介:孙文涛(1988—),男,博士,研究方向为代谢工程与合成生物学。E-mail:sunwentao2016@126.com 。 -
基金资助:国家重点研发计划(2018YFA0901800);国家自然科学基金(21736002)
Design and construction of microbial cell factory for biosynthesis of plant natural products
- Department of Chemical Engineering, Tsinghua University, Beijing 100084, China
-
Received:2020-10-20Online:2021-03-05Published:2021-03-17 -
Contact:LI Chun
摘要:
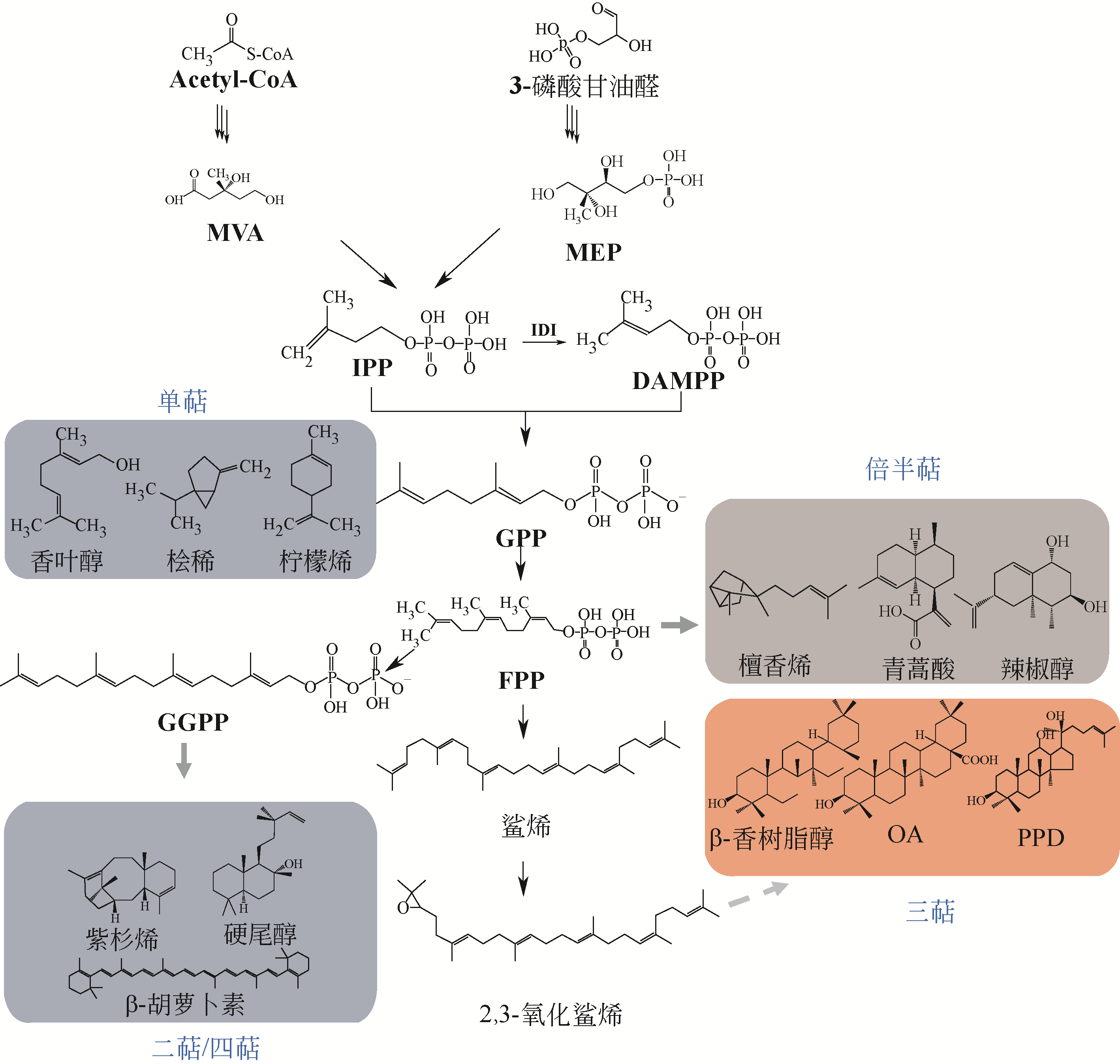
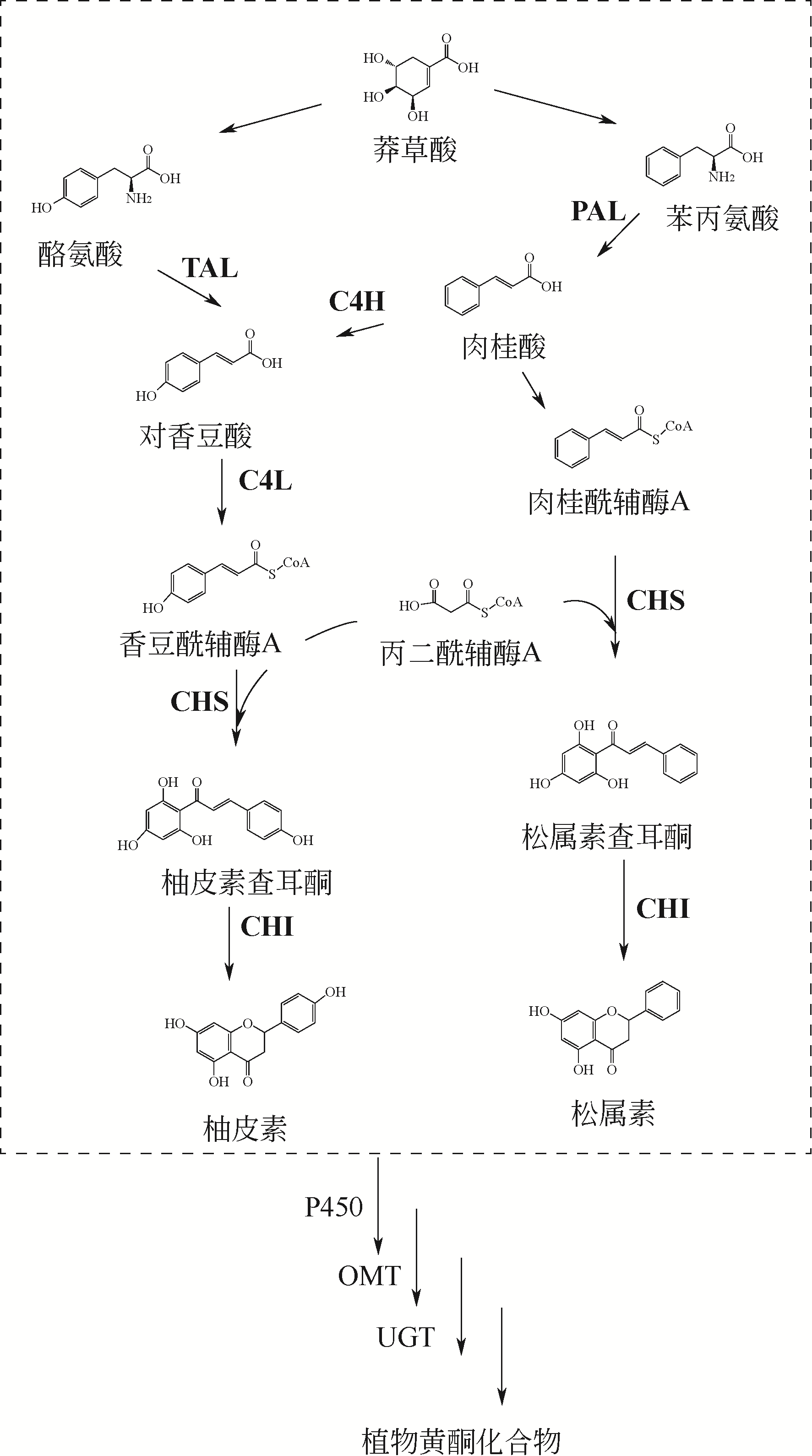
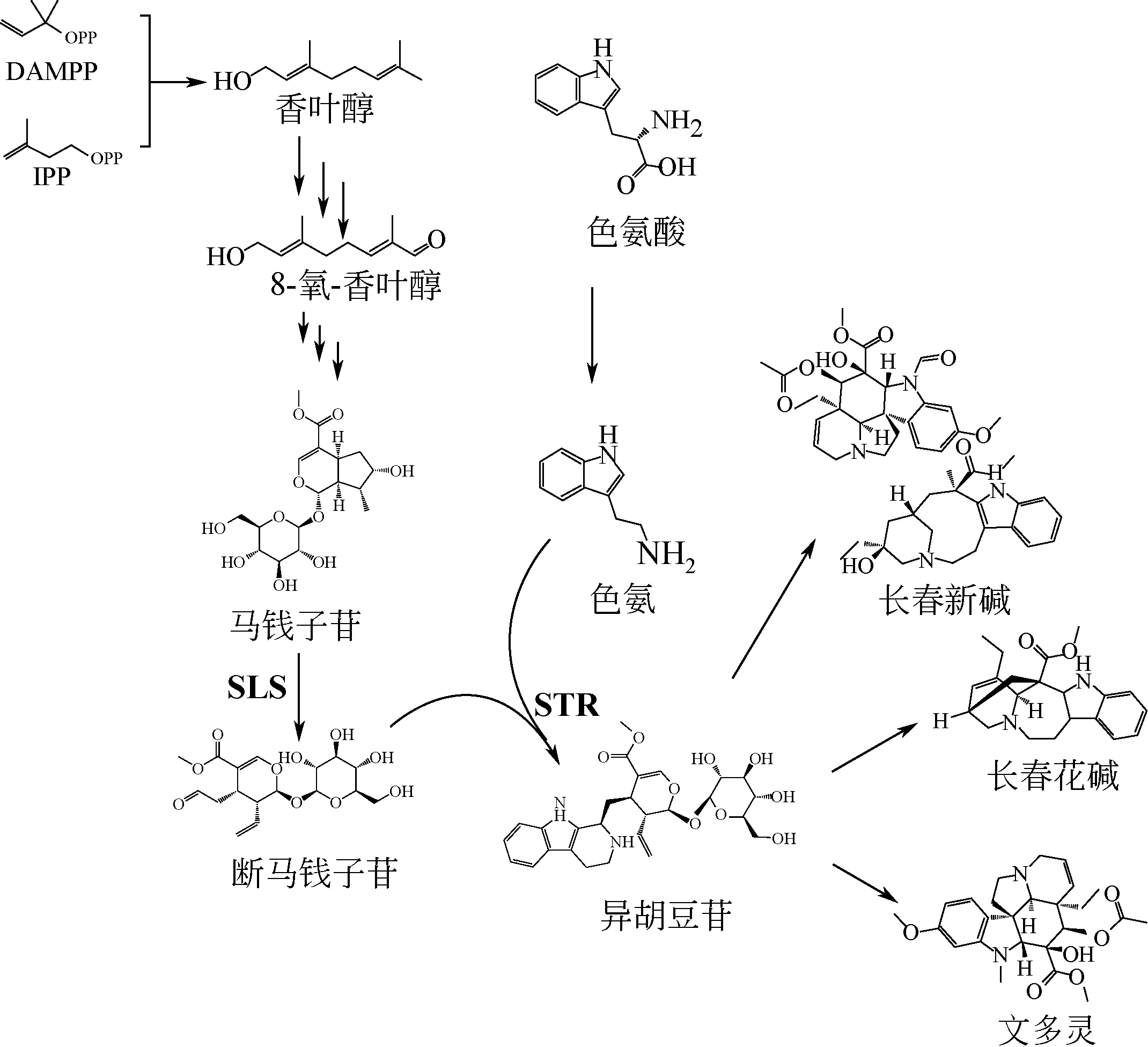
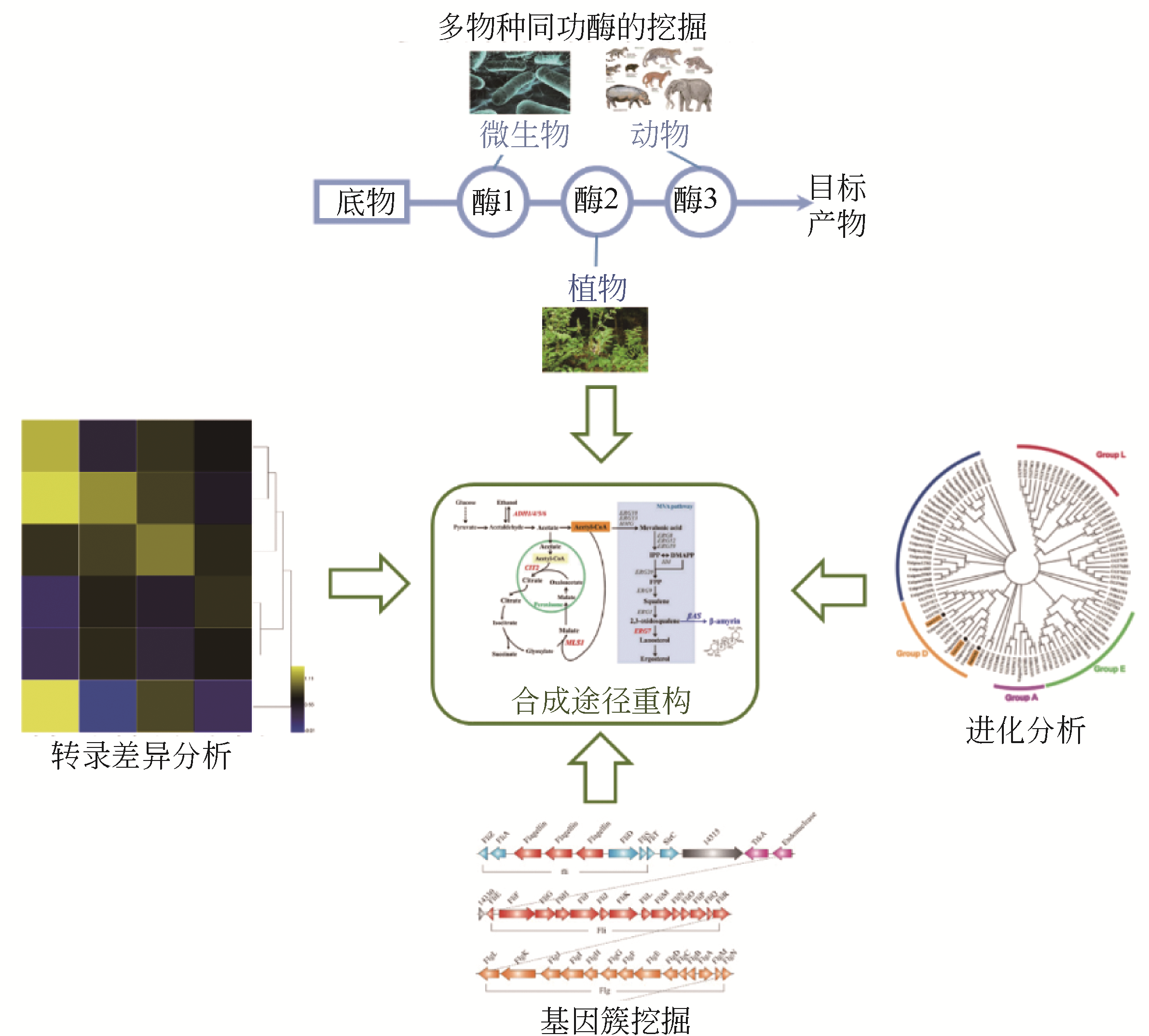
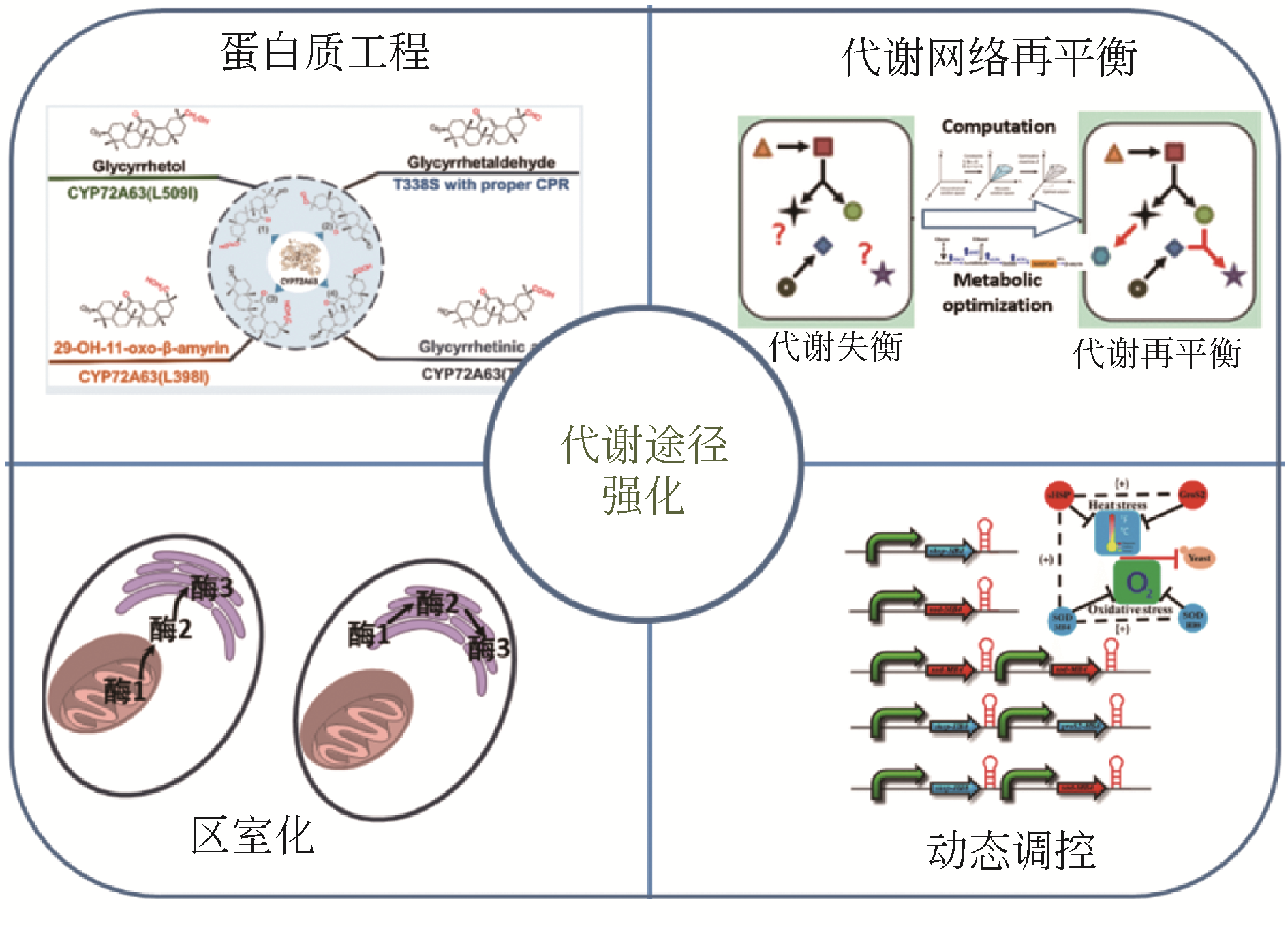
植物天然产物结构多样,具有丰富的生理活性与功能。利用微生物细胞工厂生产来源稀缺、获取难度大的植物天然产物具有经济可行、环境友好等优势。本文系统介绍了萜烯、黄酮以及生物碱的生物合成途径及其关键酶,阐述了差异转录组学、同功酶挖掘等途径解析与重构的方法。指出关键酶改造、途径动态调控、代谢区室化与代谢网络再平衡是增大外源途径代谢通量、抑制副产物合成、降低产物毒性与菌株代谢负担、提高目标产物合成能力的有效策略。提出了解析合成植物天然产物关键酶在微生物中的催化特异性机制、开发外源途径的高效组装方法等进一步提高微生物细胞工厂生产效率的建议。
中图分类号:
引用本文
孙文涛, 李春. 微生物合成植物天然产物的细胞工厂设计与构建[J]. 化工进展, 2021, 40(3): 1202-1214.
SUN Wentao, LI Chun. Design and construction of microbial cell factory for biosynthesis of plant natural products[J]. Chemical Industry and Engineering Progress, 2021, 40(3): 1202-1214.
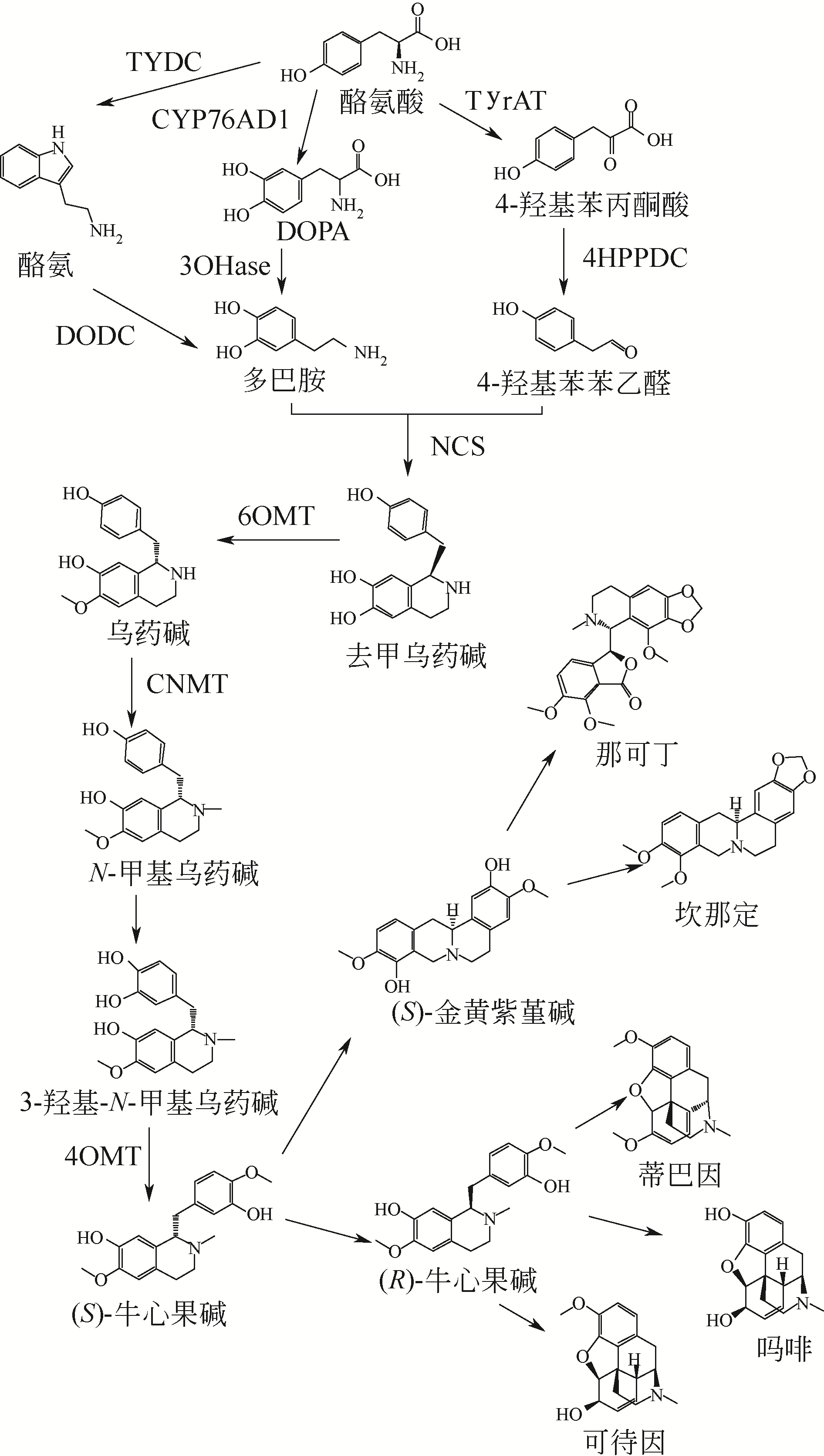
图3 苄基异喹啉生物碱合成途径TYDC—酪氨酸脱羧酶;TyrAT—酪氨酸转氨酶;DODC—多巴胺脱羧酶;3OHase—色胺羟化酶;4-HPPDC—4-羟基苯丙酮酸脱羧酶;NCS—去甲乌药碱合酶;6OMT—去甲乌药碱6-O-甲基转移酶;CNMT—乌药碱-N-甲基转移酶;4OMT—3-羟基-N-甲基-乌药碱-4-O-甲基转移酶
| 1 | KRIVORUCHKO A, NIELSEN J. Production of natural products through metabolic engineering of Saccharomyces cerevisiae[J]. Current Opinion in Biotechnology, 2015, 35: 7-15. |
| 2 | CHEMLER J A, KOFFAS M A. Metabolic engineering for plant natural product biosynthesis in microbes[J]. Current Opinion in Biotechnology, 2008, 19(6): 597-605. |
| 3 | ZHANG G L, CAO Q, LIU J Z, et al. Refactoring β-amyrin synthesis in Saccharomyces cerevisiae[J]. AIChE Journal, 2015, 61(10): 3172-3179. |
| 4 | PANDEY R P, PARAJULI P, KOFFAS M A G, et al. Microbial production of natural and non-natural flavonoids: pathway engineering, directed evolution and systems/synthetic biology[J]. Biotechnology Advances, 2016, 34(5): 634-662. |
| 5 | YU Y, CHANG P C, YU H, et al. Productive amyrin synthases for efficient alpha-amyrin synthesis in engineered Saccharomyces cerevisiae[J]. ACS Synthetic Biology, 2018, 7(10): 2391-2402. |
| 6 | SUN W T, QIN L, XUE H J, et al. Novel trends for producing plant triterpenoids in yeast[J]. Critical Reviews in Biotechnology, 2019, 39(5): 618-632. |
| 7 | XU Y H, LIU Y L, RASOOL A, et al. Sequence editing strategy for improving performance of β-glucuronidase from Aspergillus terreus[J]. Chemical Engineering Science, 2017, 167:145-153. |
| 8 | WU S, CHAPPELL J. Metabolic engineering of natural products in plants; tools of the trade and challenges for the future[J]. Current Opinion in Biotechnology, 2008, 19(2): 145-152. |
| 9 | NICOLAOU K C, YANG Z, LIU J J, et al. Total synthesis of taxol[J]. Nature, 1994, 367(6464): 630-634. |
| 10 | ZHAO Y J, LV B, FENG X D, et al. Perspective on biotransformation and De Novo biosynthesis of licorice constituents[J]. Journal of Agricultural and Food Chemistry, 2017, 65(51): 11147-11156. |
| 11 | KIM H J, TURNER T L, JIN Y S. Combinatorial genetic perturbation to refine metabolic circuits for producing biofuels and biochemicals[J]. Biotechnology Advances, 2013, 31(6): 976-985. |
| 12 | PADDON C J, WESTFALL P J, PITERA D J, et al. High-level semi-synthetic production of the potent antimalarial artemisinin[J]. Nature, 2013, 496(7446): 528-532. |
| 13 | CAO H, CHEN X Q, JASSBI A R, et al. Microbial biotransformation of bioactive flavonoids[J]. Biotechnology Advances, 2015, 33(1): 214-223. |
| 14 | SCHLAGER S, DRAGER B. Exploiting plant alkaloids[J]. Current Opinion in Biotechnology, 2016, 37:155-164. |
| 15 | O’CONNOR S E, MARESH J J. Chemistry and biology of monoterpene indole alkaloid biosynthesis[J]. Natural Product Reports, 2006, 23(4): 532-547. |
| 16 | SATO F, INUI T, TAKEMURA T. Metabolic engineering in isoquinoline alkaloid biosynthesis[J]. Current Pharmaceutical Biotechnology, 2007, 8(4): 211-218. |
| 17 | ZHANG R H, LI C Y, WANG J, et al. Microbial production of small medicinal molecules and biologics: from nature to synthetic pathways[J]. Biotechnology Advances, 2018, 36(8): 2219-2231. |
| 18 | SEKI H, SAWAI S, OHYAMA K, et al. Triterpene functional genomics in licorice for identification of CYP72A154 involved in the biosynthesis of glycyrrhizin[J]. Plant Cell, 2011, 23(11): 4112-4123. |
| 19 | SEKI H, OHYAMA K, SAWAI S, et al. Licorice β-amyrin 11-oxidase, a cytochrome P450 with a key role in the biosynthesis of the triterpene sweetener glycyrrhizin[J]. Proceedings of the National Academy of Sciences, 2008, 105(37): 14204-14209. |
| 20 | DAI Z B, LIU Y, SUN Z T, et al. Identification of a novel cytochrome P450 enzyme that catalyzes the C-2α hydroxylation of pentacyclic triterpenoids and its application in yeast cell factories[J]. Metabolic Engineering, 2019, 51: 70-78. |
| 21 | HAGEL J M, FACCHINI P J. Dioxygenases catalyze the O-demethylation steps of morphine biosynthesis in opium poppy[J]. Nature Chemical Biology, 2010, 6(4): 273-275. |
| 22 | ALLEN R S, MILLGATE A G, CHITTY J A, et al. RNAi-mediated replacement of morphine with the nonnarcotic alkaloid reticuline in opium poppy[J]. Nature Biotechnology, 2004, 22(12): 1559-1566. |
| 23 | FARROW S C, HAGEL J M, BEAUDOIN G A, et al. Stereochemical inversion of (S)-reticuline by a cytochrome P450 fusion in opium poppy[J]. Nature Biotechnology, 2015, 11(9): 728-732. |
| 24 | MEDEMA M H, OSBOURN A. Computational genomic identification and functional reconstitution of plant natural product biosynthetic pathways[J]. Natural Product Reports, 2016, 33(8): 951-962. |
| 25 | FIELD B, OSBOURN A E. Metabolic diversification—independent assembly of operon-like gene clusters in different plants[J]. Science, 2008, 320(5875): 543-547. |
| 26 | FIELD B, A-S FISTON-LAVIER, KEMEN A, et al. Formation of plant metabolic gene clusters within dynamic chromosomal regions[J]. Proceedings of the National Academy of Sciences of the United States of America, 2011, 108(38): 16116-16121. |
| 27 | WINZER T, GAZDA V, HE Z, et al. A Papaver somniferum 10-gene cluster for synthesis of the anticancer alkaloid noscapine[J]. Science, 2012, 336(6089): 1704-1708. |
| 28 | BROWN S F, BRANFORD A J, MORAN W. On the use of artificial neural networks for the analysis of survival data[J]. IEEE Transactions On Neural Networks, 1997, 8(5): 1071-1077. |
| 29 | KAUTSAR S A, SUAREZ DURAN H G, BLIN K, et al. PlantiSMASH: automated identification, annotation and expression analysis of plant biosynthetic gene clusters[J]. Nucleic Acids Research, 2017, 45(W1): W55-W63. |
| 30 | WEBER T, BLIN K, DUDDELA S, et al. AntiSMASH 3.0-a comprehensive resource for the genome mining of biosynthetic gene clusters[J]. Nucleic Acids Research, 2015, 43(1): W237-W243. |
| 31 | DENOEUD F, CARRETERO-PAULET L, DEREEPER A, et al. The coffee genome provides insight into the convergent evolution of caffeine biosynthesis[J]. Science, 2014, 345(6201): 1181-1184. |
| 32 | ZHU M, WANG C X, SUN W T, et al. Boosting 11-oxo-beta-amyrin and glycyrrhetinic acid synthesis in Saccharomyces cerevisiae via pairing novel oxidation and reduction system from legume plants[J]. Metabolic Engineering, 2018, 45: 43-50. |
| 33 | HWANG E I, KANEKO M, OHNISHI Y, et al. Production of plant-specific flavanones by Escherichia coli containing an artificial gene cluster[J]. Applied and Environmental Microbiology, 2003, 69(5): 2699-2706. |
| 34 | TRENCHARD I J, SIDDIQUI M S, THODEY K, et al. De novo production of the key branch point benzylisoquinoline alkaloid reticuline in yeast[J]. Metabolic Engineering, 2015, 31:74-83. |
| 35 | NAKAGAWA A, MINAMI H, KIM J S, et al. A bacterial platform for fermentative production of plant alkaloids[J]. Nature Communications, 2011, 2(1): 326. |
| 36 | SIDDIQUI M S, KATE T, ISIS T, et al. Advancing secondary metabolite biosynthesis in yeast with synthetic biology tools[J]. FEMS Yeast Research, 2012, 12(2): 144-170. |
| 37 | HWANG E I, KANEKO M, OHNISHI Y, et al. Production of plant-specific flavanones by Escherichia coli containing an artificial gene cluster[J]. Applied & Environmental Microbiology, 2003, 69(5): 2699-2706. |
| 38 | DELOACHE W C, RUSS Z N, NARCROSS L, et al. An enzyme-coupled biosensor enables (S)-reticuline production in yeast from glucose[J]. Nature Chemical Biology, 2015, 11(7): 465-471. |
| 39 | DIETRICH J A, YOSHIKUNI Y, FISHER K, et al. A novel semi-biosynthetic route for artemisinin production using engineered substrate-promiscuous P450BM3[J]. ACS Chemical Biology, 2009, 4(4): 261-267. |
| 40 | CHANG M C Y, EACHUS R A, TRIEU W, et al. Engineering Escherichia coli for production of functionalized terpenoids using plant P450s[J]. Nature Chemical Biology, 2007, 3(5): 274-277. |
| 41 | SUN W T, XUE H J, LIU H, et al. Controlling chemo- and regioselectivity of a plant P450 in yeast cell toward rare licorice triterpenoid biosynthesis[J]. ACS Catalysis, 2020,10: 4253-4260. |
| 42 | DELOACHE W C, RUSS Z N, NARCROSS L, et al. An enzyme-coupled biosensor enables (S)-reticuline production in yeast from glucose[J]. Nature Chemical Biology, 2015, 11(7): 465-471. |
| 43 | XIE W P, LV X M, YE L, et al. Construction of lycopene-overproducing Saccharomyces cerevisiae by combining directed evolution and metabolic engineering[J]. Metabolic Engineering, 2015, 30:69-78. |
| 44 | WANG P P, WEI W, YE W, et al. Synthesizing ginsenoside Rh2 in Saccharomyces cerevisiae cell factory at high-efficiency[J]. Cell Discovery, 2019, 5(1): 1-14. |
| 45 | EDGAR S, LI F S, QIAO K, et al. Engineering of taxadiene synthase for improved selectivity and yield of a key taxol biosynthetic intermediate[J]. ACS Synthetic Biology, 2016, 6(2): 201-205. |
| 46 | XIONG S T, WANG Y, YAO M D, et al. Cell foundry with high product specificity and catalytic activity for 21-deoxycortisol biotransformation[J]. Microbial Cell Factories, 2017, 16(1): 105. |
| 47 | GONZALEZ F J, KORZEKWA K R. Cytochromes P450 expression systems[J]. Annual Review of Pharmacology & Toxicology, 1995, 35(1): 369-390. |
| 48 | BIGGS B W, LIM C G, SAGLIANI K, et al. Overcoming heterologous protein interdependency to optimize P450-mediated Taxol precursor synthesis in Escherichia coli[J]. Proceedings of the National Academy of Sciences of the United States of America, 2016, 113(12): 3209-3214. |
| 49 | MUNRO A W, GIRVAN H M, MCLEAN K J. Variations on a (t)heme—novel mechanisms, redox partners and catalytic functions in the cytochrome P450 superfamily[J]. Natural Product Reports, 2007, 24(3): 585-609. |
| 50 | GALANIE S, THODEY K, TRENCHARD I J, et al. Complete biosynthesis of opioids in yeast[J]. Science, 2015, 349(6252): 1095-1100. |
| 51 | SOH K C, HATZIMANIKATIS V. Dreams of metabolism[J]. Trends in Biotechnology, 2010, 28(10): 501-508. |
| 52 | RENAULT H, BASSARD J E, HAMBERGER B, et al. Cytochrome P450-mediated metabolic engineering: current progress and future challenges[J]. Current Opinion in Plant Biology, 2014, 19: 27-34. |
| 53 | PADDON C J, KEASLING J D. Semi-synthetic artemisinin: a model for the use of synthetic biology in pharmaceutical development[J]. Nature Reviews Microbiology, 2014, 12(5): 355-367. |
| 54 | YANG D, PARK S Y, PARK Y S, et al. Metabolic engineering of Escherichia coli for natural product biosynthesis[J]. Trends in Biotechnology, 2020, 38(7):745-765. |
| 55 | ZHAO Y J, FAN J J, WANG C, et al. Enhancing oleanolic acid production in engineered Saccharomyces cerevisiae[J]. Bioresource Technology, 2018, 257:339-343. |
| 56 | LIAO P, HEMMERLIN A, BACH T J, et al. The potential of the mevalonate pathway for enhanced isoprenoid production[J]. Biotechnology Advances, 2016, 34(5): 697-713. |
| 57 | LIANG C N, ZHANG X X, WU J Y, et al. Dynamic control of toxic natural product biosynthesis by an artificial regulatory circuit[J]. Metabolic Engineering, 2020, 57: 239-246. |
| 58 | SCALCINATI G, KNUF C, PARTOW S, et al. Dynamic control of gene expression in Saccharomyces cerevisiae engineered for the production of plant sesquitepene alpha-santalene in a fed-batch mode[J]. Metabolic Engineering, 2012, 14(2): 91-103. |
| 59 | SKJOEDT M L, SNOEK T, KILDEGAARD K R, et al. Engineering prokaryotic transcriptional activators as metabolite biosensors in yeast[J]. Nature Chemical Biology, 2016, 12(11): 951-958. |
| 60 | SIEDLER S, STAHLHUT S G, MALLA S, et al. Novel biosensors based on flavonoid-responsive transcriptional regulators introduced into Escherichia coli[J]. Metabolic Engineering, 2014, 21: 2-8. |
| 61 | LIU D, XIAO Y, EVANS B S, et al. Negative feedback regulation of fatty acid production based on a malonyl-CoA sensor-actuator[J]. ACS Synthetic Biology, 2015, 4(2): 132-140. |
| 62 | XU P, WANG W Y, LI L Y, et al. Design and kinetic analysis of a hybrid promoter-regulator system for malonyl-CoA sensing in Escherichia coli[J]. ACS Chemical Biology, 2013, 9(2): 451-458. |
| 63 | LANGAN R A, BOYKEN S E, NG A H, et al. De novo design of bioactive protein switches[J]. Nature, 2019, 572:205-210. |
| 64 | ZHAO E M, ZHANG Y, MEHL J, et al. Optogenetic regulation of engineered cellular metabolism for microbial chemical production[J]. Nature, 2018, 555(7698): 683-687. |
| 65 | QIN J F, ZHOU Y J, KRIVORUCHKO A, et al. Modular pathway rewiring of Saccharomyces cerevisiae enables high-level production of L-ornithine[J]. Nature Communications, 2015, 6(1): 1-11. |
| 66 | ZHAO S, JONES J A, LACHANCE D M, et al. Improvement of catechin production in Escherichia coli through combinatorial metabolic engineering[J]. Metabolic Engineering, 2015, 28: 43-53. |
| 67 | GAIRIK S, ABHISHEK G, DAVID G, et al. In vivo co-localization of enzymes on RNA scaffolds increases metabolic production in a geometrically dependent manner[J]. Nucleic Acids Research, 2014, 42(14):9493-9503. |
| 68 | BROWN S, CLASTRE M, COURDAVAULT V, et al. De novo production of the plant-derived alkaloid strictosidine in yeast[J]. Proceedings of the National Academy of Sciences of the United States of America, 2015, 112(11): 3205-3210. |
| 69 | DAI Z J, HUANG M T, CHEN Y, et al. Global rewiring of cellular metabolism renders Saccharomyces cerevisiae Crabtree negative[J]. Nature Communications, 2018, 9(1): 3059. |
| 70 | MITCHELL L A, WANG A, STRACQUADANIO G, et al. Synthesis, debugging, and effects of synthetic chromosome consolidation: synⅥ and beyond[J]. Science, 2017, 355(6329): eaaf4831. |
| 71 | SHEN Y, WANG Y, CHEN T, et al. Deep functional analysis of synⅡ, a 770-kilobase synthetic yeast chromosome[J]. Science, 2017, 355(6329):eaaf4791. |
| 72 | ZHOU K, QIAO K J, EDGAR S, et al. Distributing a metabolic pathway among a microbial consortium enhances production of natural products[J]. Nature Biotechnology, 2015, 33(4): 377-383. |
| 73 | JONES J A, VERNACCHIO V R, SINKOE A L, et al. Experimental and computational optimization of an Escherichia coli co-culture for the efficient production of flavonoids[J]. Metabolic Engineering, 2016, 35: 55-63. |
| 74 | JONES J A, VERNACCHIO V R, COLLINS S M, et al. Complete biosynthesis of anthocyanins using E. coli polycultures[J]. mBio, 2017, 8(3): e00621-e00617. |
| [1] | 高聪, 陈城虎, 陈修来, 刘立明. 代谢工程改造微生物合成生物基单体的进展与挑战[J]. 化工进展, 2023, 42(8): 4123-4135. |
| [2] | 郭峰, 张尚杰, 蒋羽佳, 姜万奎, 信丰学, 章文明, 姜岷. 一碳资源在酵母中的利用与转化[J]. 化工进展, 2023, 42(1): 30-39. |
| [3] | 陶雨萱, 张尚杰, 景艺文, 信丰学, 董维亮, 周杰, 蒋羽佳, 章文明, 姜岷. 甲基营养型大肠杆菌构建策略的研究进展[J]. 化工进展, 2021, 40(7): 3932-3941. |
| [4] | 郭亮, 高聪, 张丽, 陈修来, 刘立明. 人工代谢路径适配性的研究进展[J]. 化工进展, 2021, 40(3): 1252-1261. |
| [5] | 王颖, 曲俊泽, 梁楠, 郝鹤, 元英进. 合成类胡萝卜素细胞工厂的快速构建和定向进化[J]. 化工进展, 2021, 40(3): 1187-1201. |
| [6] | 刘卫兵, 叶邦策. 放线菌聚酮类化合物的合成生物学研究及生物制造[J]. 化工进展, 2021, 40(3): 1226-1237. |
| [7] | 马悦原, 陈金春, 陈国强. 嗜盐微生物底盘细胞:应用和前景[J]. 化工进展, 2021, 40(3): 1178-1186. |
| [8] | 高聪, 郭亮, 胡贵鹏, 陈修来, 刘立明. 微生物细胞工厂碳流调控进展[J]. 化工进展, 2021, 40(12): 6807-6817. |
| [9] | 王琛, 赵猛, 丁明珠, 王颖, 姚明东, 肖文海. 生物支架系统在合成生物学中的应用[J]. 化工进展, 2020, 39(11): 4557-4567. |
| [10] | 常鹏程, 于洋, 王颖, 李春. 酿酒酵母高效合成萜类化合物的组合调控策略[J]. 化工进展, 2019, 38(01): 598-605. |
| [11] | 张正晖, 曹铭铭, 李珺, 李春, 刘护. 微生物高效分泌蛋白质的策略与应用[J]. 化工进展, 2018, 37(08): 3129-3137. |
| [12] | 樊婧婧, 赵雨佳, 王晨, 李春, 周晓宏. 酿酒酵母乙酰辅酶A精细调控合成萜类化合物研究进展[J]. 化工进展, 2018, 37(07): 2773-2779. |
| [13] | 杨坤, 王颖, 李春. 细胞转运蛋白及其工程化应用[J]. 化工进展, 2017, 36(04): 1410-1417. |
| [14] | 刘丁玉, 孟娇, 王智文, 陈涛, 赵学明. 多元模块工程在代谢工程中的应用与研究进展[J]. 化工进展, 2016, 35(11): 3619-3626. |
| [15] | 肖文海, 周嗣杰, 王颖, 元英进. 如何工程化生物学[J]. 化工进展, 2016, 35(06): 1827-1836. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||