化工进展 ›› 2021, Vol. 40 ›› Issue (3): 1226-1237.DOI: 10.16085/j.issn.1000-6613.2020-1906
放线菌聚酮类化合物的合成生物学研究及生物制造
- 华东理工大学微分析与生物系统工程实验室,生物反应器工程国家重点实验室,上海 200237
-
收稿日期:2020-09-19出版日期:2021-03-05发布日期:2021-03-17 -
通讯作者:叶邦策 -
作者简介:刘卫兵(1978—),男,博士,副教授,研究方向为合成生物学与微生物代谢调控。E-mail:lwb@ecust.edu.cn 。 -
基金资助:国家自然科学基金(31730004);国家重点研发计划“合成生物学”重点专项(2018YFA0900404);上海市自然科学基金面上项目(19ZR1413700)
Progress of synthetic biology research and biological manufacturing of actinomycetes polyketides
- Laboratory of Biosystems and Microanalysis, State Key Laboratory of Bioreactor Engineering, East China University of Science and Technology, Shanghai 200237, China
-
Received:2020-09-19Online:2021-03-05Published:2021-03-17 -
Contact:YE Bangce
摘要:
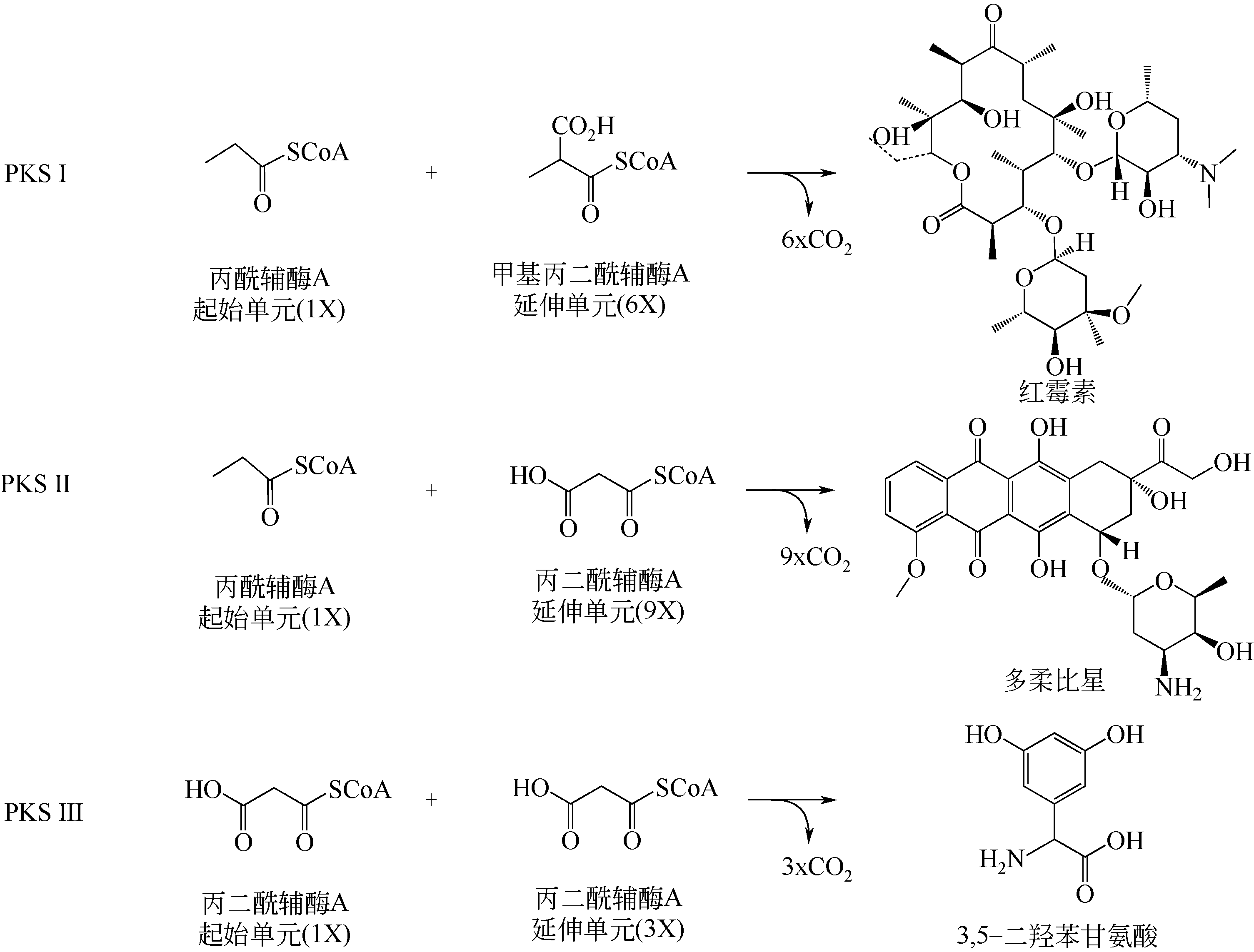
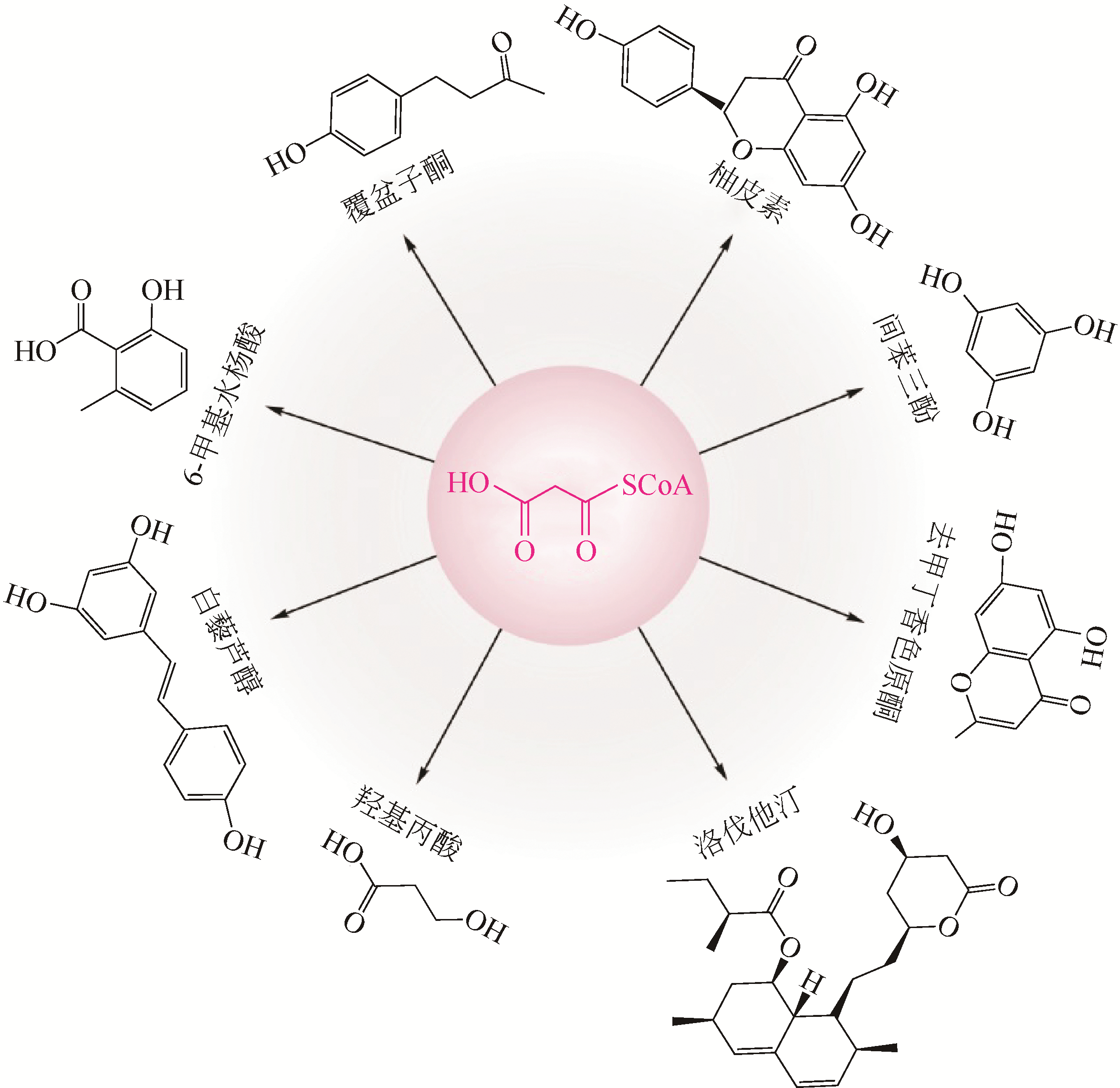
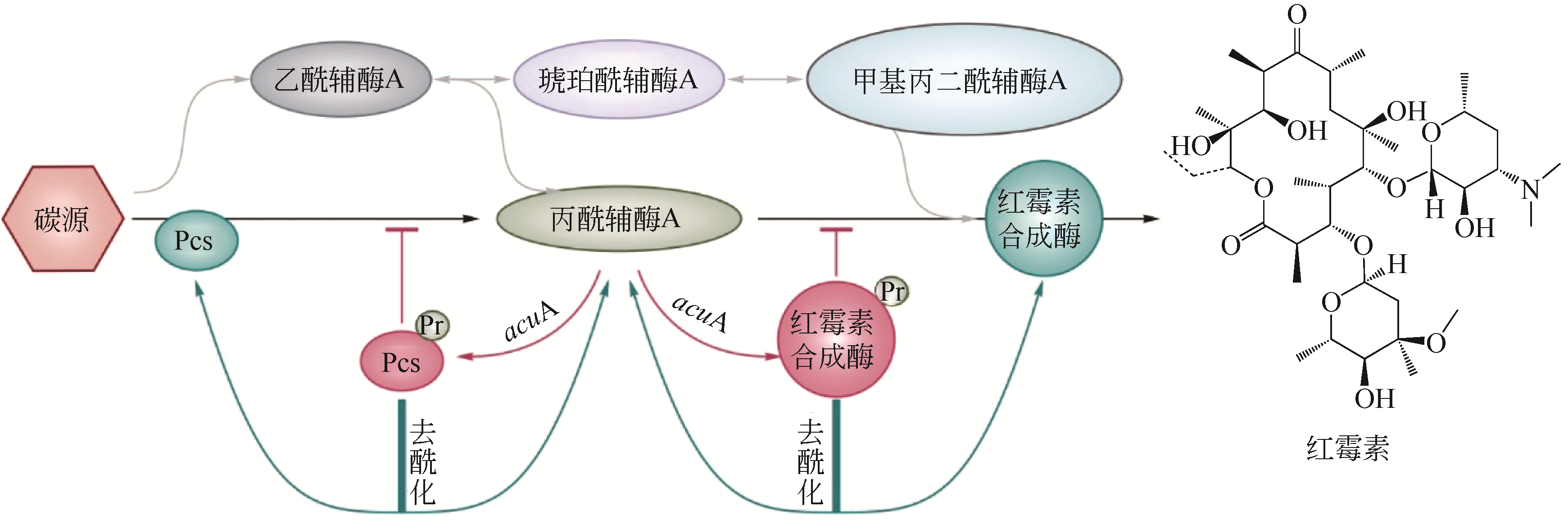
聚酮化合物具有广泛的药用活性和极高的经济价值,但如何高效、经济、绿色、环保地合成聚酮化合物是目前急需解决的问题。随着合成生物学的发展及分子生物学技术的进步,不断有新的技术和策略被用于聚酮化合物的生物制造。本文介绍了聚酮化合物生物制造中的关键酶、前体物质及代谢途径等,分析了通过CRISPR技术及翻译后修饰代谢工程优化代谢调控网络;通过替换及优化启动子等手段改造与优化代谢途径;通过构建简单、高效的异源表达系统等策略提高聚酮化合物的生物制造效率等。在此基础上对红霉素、阿维菌素、多杀菌素的合成生物学研究的最新进展进行了总结,进而对当前聚酮化合物生物制造面临的产量及效率低下等问题和可能的解决途径,如平衡初级代谢与次级代谢,构建新型、优势底盘细胞及代谢网络的重新设计与改造等进行了展望。
中图分类号:
引用本文
刘卫兵, 叶邦策. 放线菌聚酮类化合物的合成生物学研究及生物制造[J]. 化工进展, 2021, 40(3): 1226-1237.
LIU Weibing, YE Bangce. Progress of synthetic biology research and biological manufacturing of actinomycetes polyketides[J]. Chemical Industry and Engineering Progress, 2021, 40(3): 1226-1237.
| 1 | HERTWECK C. The biosynthetic logic of polyketide diversity[J]. Angewandte Chemie: International Edition, 2009, 48(26): 4688-4716. |
| 2 | MA S M, LI J W H, CHOI J W, et al. Complete reconstitution of a highly reducing iterative polyketide synthase[J]. Science, 2009, 326(5952): 589-592. |
| 3 | HLEBA L, CHAROUSOVA I, CISAROVA M, et al. Rapid identification of Streptomyces tetracycline producers by MALDI-TOF mass spectrometry[J]. Journal of Environmental Science and Health: Part A, 2018, 53(12): 1083-1093. |
| 4 | PARTHASARATHY R, SATHIYABAMA M. Lovastatin-producing endophytic fungus isolated from a medicinal plant Solanum xanthocarpum[J]. Natural Product Research, 2015, 29(24): 2282-2286. |
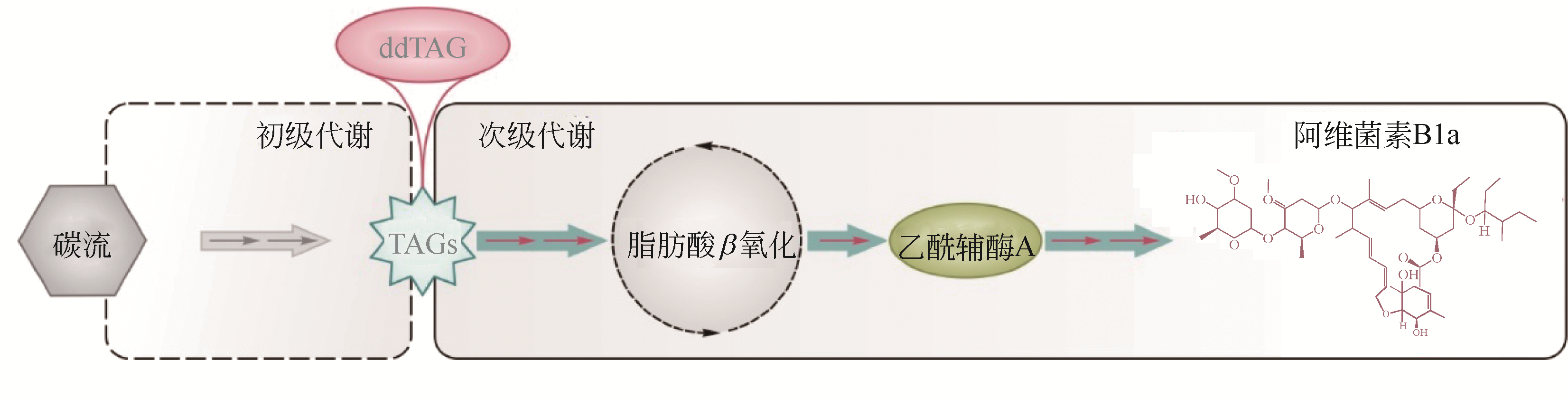
| 5 | HUANG Q, LU G, SHEN H M, et al. Anti-cancer properties of anthraquinones from rhubarb[J]. Medicinal Research Reviews, 2007, 27(5): 609-630. |
| 6 | PANKEWITZ F, HILKER M. Polyketides in insects: ecological role of these widespread chemicals and evolutionary aspects of their biogenesis[J]. Biological Reviews, 2008, 83(2): 209-226. |
| 7 | KATSUYAMA Y, FUNA N, MIYAHISA I, et al. Synthesis of unnatural flavonoids and stilbenes by exploiting the plant biosynthetic pathway in Escherichia coli[J]. Chemistry & Biology, 2007, 14(6): 613-621. |
| 8 | CHOPRA I, ROBERTS M. Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance[J]. Microbiology and Molecular Biology Reviews, 2001, 65(2): 232-260. |
| 9 | GHANNOUM M A, RICE L B. Antifungal agents: mode of action, mechanisms of resistance, and correlation of these mechanisms with bacterial resistance[J]. Clinical Microbiology Reviews, 1999, 12(4): 501-517. |
| 10 | TACAR O, SRIAMORNSAK P, DASS C R. Doxorubicin: an update on anticancer molecular action, toxicity and novel drug delivery systems[J]. Journal of Pharmacy and Pharmacology, 2013, 65(2): 157-170. |
| 11 | SHUSHNI M A, SINGH R, MENTEL R, et al. Balticolid: a new 12-membered macrolide with antiviral activity from an ascomycetous fungus of marine origin[J]. Marine Drugs, 2011, 9(5): 844-851. |
| 12 | LI J, KIM S G, BLENIS J. Rapamycin: one drug, many effects[J]. Cell Metabolism, 2014, 19(3): 373-379. |
| 13 | DE DONK N W VAN, KAMPHUIS M M, LOKHORST H M, et al. The cholesterol lowering drug lovastatin induces cell death in myeloma plasma cells[J]. Leukemia, 2002, 16(7): 1362-1371. |
| 14 | NEWMAN D J, CRAGG G M. Natural products as sources of new drugs over the nearly four decades from 01/1981 to 09/2019[J]. Journal of Natural Products, 2020, 83(3): 770-803. |
| 15 | GOMES E S, SCHUCH V, LEMOS E G D M. Biotechnology of polyketides: new breath of life for the novel antibiotic genetic pathways discovery through metagenomics[J]. Brazilian Journal of Microbiology, 2013, 44(4): 1007-1034. |
| 78 | MARKHAM K A, PALMER C M, CHWATKO M, et al. Rewiring Yarrowia lipolytica toward triacetic acid lactone for materials generation[J]. Proceedings of the National Academy of Sciences of the United States of America, 2018, 115(9): 2096-2101. |
| 79 | TAN Z G, CLOMBURG J M, CHEONG S, et al. A polyketoacyl-CoA thiolase-dependent pathway for the synthesis of polyketide backbones[J]. Nature Catalysis, 2020, 3(7): 593. |
| 80 | RISDIAN C, MOZEF T, WINK J. Biosynthesis of polyketides in Streptomyces[J]. Microorganisms, 2019, 7(5): 124. |
| 81 | LEADLAY P F. Combinatorial approaches to polyketide biosynthesis[J]. Curr. Opin. Chem. Biol., 1997, 1(2): 162-168. |
| 82 | XU Z, WANG M, YE B C. TetR family transcriptional regulator PccD negatively controls propionyl coenzyme A assimilation in Saccharopolyspora erythraea[J]. J. Bacteriol., 2017, 199(20): e00281. |
| 83 | XU J Y, XU Z, ZHOU Y, et al. Lysine malonylome may affect the central metabolism and erythromycin biosynthesis pathway in Saccharopolyspora erythraea[J]. J. Proteome. Res., 2016, 15(5): 1685-1701. |
| 84 | LYU M, CHENG Y, HAN X, et al. AccR, a TetR family transcriptional repressor, coordinates short-chain acyl coenzyme a homeostasis in Streptomyces avermitilis[J]. Appl. Environ. Microbiol., 2020, 86(12): e00508-e00520. |
| 85 | LU X, LIU X, CHEN Z, et al. The ROK-family regulator Rok7B7 directly controls carbon catabolite repression, antibiotic biosynthesis, and morphological development in Streptomyces avermitilis[J]. Environ. Microbiol., 2020, 22(12): 5090-5108. |
| 86 | ZHAO C, HUANG Y, GUO C, et al. Heterologous expression of spinosyn biosynthetic gene cluster in streptomyces species is dependent on the expression of rhamnose biosynthesis genes[J]. J. Mol. Microbiol. Biotechnol., 2017, 27(3): 190-198. |
| 87 | TAN G Y, DENG K, LIU X, et al. Heterologous biosynthesis of spinosad: an omics-guided large polyketide synthase gene cluster reconstitution in streptomyces[J]. ACS Synth. Biol., 2017, 6(6): 995-1005. |
| 16 | CHENG S, LIU X, JIANG G, et al. Orthogonal engineering of biosynthetic pathway for efficient production of limonene in Saccharomyces cerevisiae[J]. ACS Synth. Biol., 2019, 8(5): 968-975. |
| 17 | ITOH H, MIURA A, MATSUI M, et al. Knockout of the SREBP system increases production of the polyketide FR901512 in filamentous fungal sp. No. 14919 and lovastatin in Aspergillus terreus ATCC20542[J]. Appl. Microbiol. Biotechnol., 2018, 102(3): 1393-1405. |
| 18 | XU Y, LI Y X, YE B C. Lysine propionylation modulates the transcriptional activity of phosphate regulator PhoP in Saccharopolyspora erythraea[J]. Mol. Microbiol., 2018, 110(4): 648-661. |
| 19 | QIN R, SANG Y, REN J, et al. The bacterial two-hybrid system uncovers the involvement of acetylation in regulating of Lrp activity in Salmonella typhimurium[J]. Front Microbiol., 2016(7): 1864. |
| 20 | XU J Y, YOU D, LENG P Q, et al. Allosteric regulation of a protein acetyltransferase in Micromonospora aurantiaca by the amino acids cysteine and arginine[J]. Journal of Biological Chemistry, 2014, 289(39): 27034-27045. |
| 21 | LIU X X, SHEN M J, LIU W B, et al. GlnR-mediated regulation of short-chain fatty acid assimilation in Mycobacterium smegmatis[J]. Frontiers in Microbiology, 2018, 9: 1311. |
| 22 | YU B J, KIM J A, MOON J H, et al. The diversity of lysine-acetylated proteins in Escherichia coli[J]. Journal of Microbiology and Biotechnology, 2008, 18(9): 1529. |
| 23 | WANG W, LI S, LI Z, et al. Harnessing the intracellular triacylglycerols for titer improvement of polyketides in Streptomyces[J]. Nature Biotechnology, 2020, 38(1): 76-83. |
| 24 | BIRCH A, DONOVAN F. Studies in relation to biosynthesis. Ⅰ. Some possible routes to derivatives of orcinol and phloroglucinol[J]. Australian Journal of Chemistry, 1953, 6(4): 360-368. |
| 25 | CORTES J, HAYDOCK S F, ROBERTS G A, et al. An unusually large multifunctional polypeptide in the erythromycin-producing polyketide synthase of Saccharopolyspora erythraea[J]. Nature, 1990, 348(6297): 176-178. |
| 26 | HOPWOOD D A, SHERMAN D H. Molecular genetics of polyketides and its comparison to fatty acid biosynthesis[J]. Annual Review of Genetics, 1990, 24(1): 37-66. |
| 27 | KATZ L, DONADIO S. Polyketide synthesis: prospects for hybrid antibiotics[J]. Annu. Rev. Microbiol., 1993, 47: 875-912. |
| 28 | CORTES J, HAYDOCK S F, ROBERTS G A, et al. An unusually large multifunctional polypeptide in the erythromycin-producing polyketide synthase of Saccharopolyspora erythraea[J]. Nature, 1990, 348(6297): 176-178. |
| 29 | DONADIO S, STAVER M J, MCALPINE J B, et al. Modular organization of genes required for complex polyketide biosynthesis[J]. Science, 1991, 252(5006): 675-679. |
| 30 | MALPARTIDA F, HOPWOOD D A. Molecular cloning of the whole biosynthetic pathway of a Streptomyces antibiotic and its expression in a heterologous host[J]. Nature, 1984, 309(5967): 462-464. |
| 31 | MOTAMEDI H, HUTCHINSON C R. Cloning and heterologous expression of a gene cluster for the biosynthesis of tetracenomycin C, the anthracycline antitumor antibiotic of Streptomyces glaucescens[J]. Proceedings of the National Academy of Sciences of the United States of America, 1987, 84(13): 4445-4449. |
| 32 | FUNA N, OHNISHI Y, FUJII I, et al. A new pathway for polyketide synthesis in microorganisms[J]. Nature, 1999, 400(6747): 897-899. |
| 33 | IKEDA H, NONOMIYA T, USAMI M, et al. Organization of the biosynthetic gene cluster for the polyketide anthelmintic macrolide avermectin in Streptomyces avermitilis[J]. Proceedings of the National Academy of Sciences of the United States of America, 1999, 96(17): 9509-9514. |
| 34 | CHAN Y A, PODEVELS A M, KEVANY B M, et al. Biosynthesis of polyketide synthase extender units[J]. Nat. Prod. Rep., 2009, 26(1): 90-114. |
| 35 | LIM Y P, GO M K, YEW W S. Exploiting the biosynthetic potential of type Ⅲ polyketide synthases[J]. Molecules, 2016, 21(6): 806. |
| 36 | RAY L, MOORE B S. Recent advances in the biosynthesis of unusual polyketide synthase substrates[J]. Nat. Prod. Rep., 2016, 33(2): 150-161. |
| 37 | JESCHEK M, GERNGROSS D, PANKE S. Combinatorial pathway optimization for streamlined metabolic engineering[J]. Curr. Opin. Biotechnol., 2017, 47: 142-151. |
| 38 | LI S, SI T, WANG M, et al. Development of a synthetic malonyl-CoA sensor in Saccharomyces cerevisiae for intracellular metabolite monitoring and genetic screening[J]. ACS Synth. Biol., 2015, 4(12): 1308-1315. |
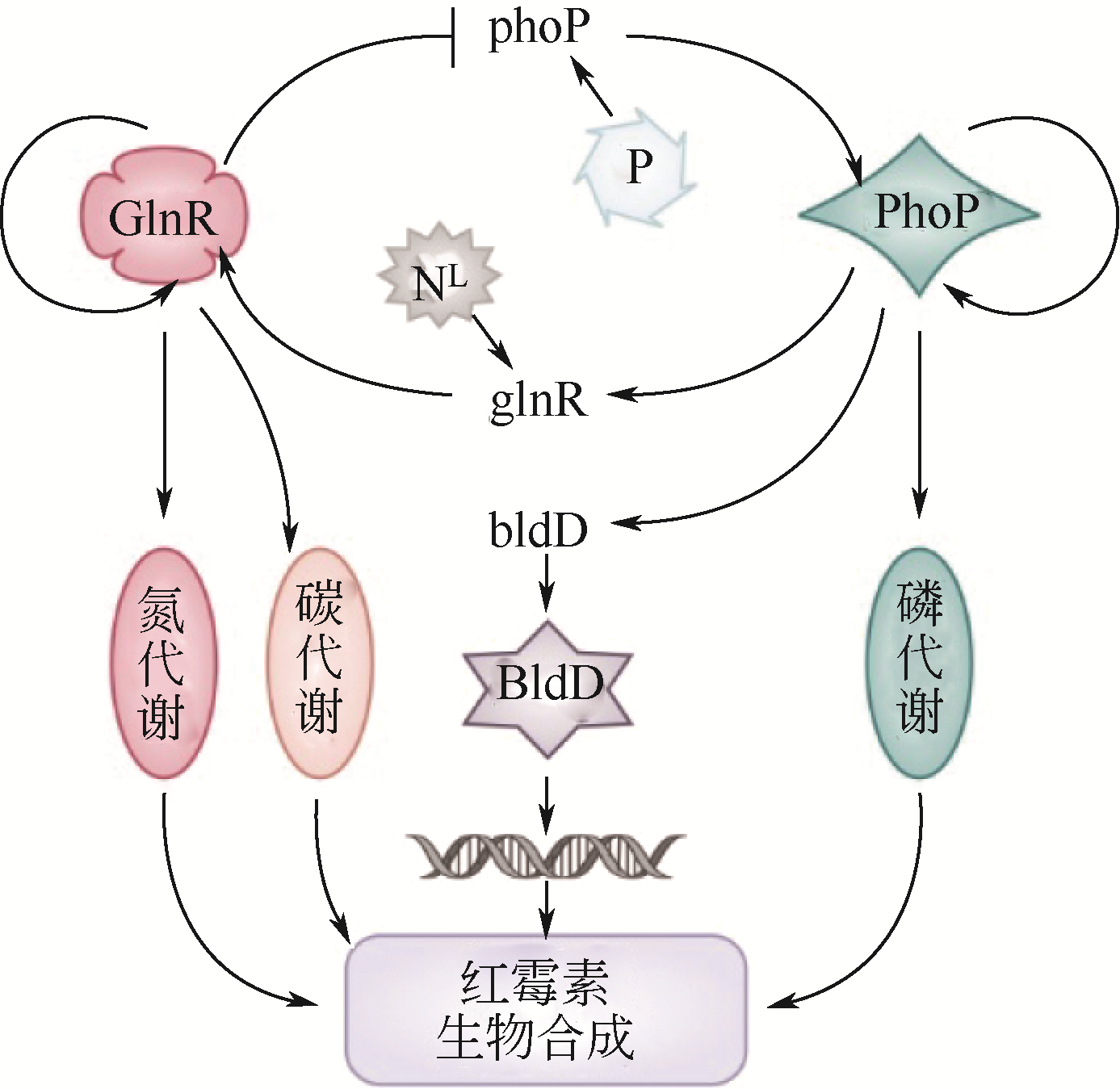
| 39 | PITERA D J, PADDON C J, NEWMAN J D, et al. Balancing a heterologous mevalonate pathway for improved isoprenoid production in Escherichia coli[J]. Metab. Eng., 2007, 9(2): 193-207. |
| 40 | NIELSEN J, KEASLING J D. Engineering cellular metabolism[J]. Cell, 2016, 164(6): 1185-1197. |
| 41 | LIM C G, FOWLER Z L, HUELLER T, et al. High-yield resveratrol production in engineered Escherichia coli[J]. Appl. Environ. Microbiol., 2011, 77(10): 3451-3460. |
| 42 | SANTOS C N, KOFFAS M, STEPHANOPOULOS G. Optimization of a heterologous pathway for the production of flavonoids from glucose[J]. Metab. Eng., 2011, 13(4): 392-400. |
| 43 | SUMMEREN-WESENHAGEN P V VAN, MARIENHAGEN J. Metabolic engineering of Escherichia coli for the synthesis of the plant polyphenol pinosylvin[J]. Appl. Environ. Microbiol., 2015, 81(3): 840-849. |
| 44 | KALLSCHEUER N, VOGT M, STENZEL A, et al. Construction of a Corynebacterium glutamicum platform strain for the production of stilbenes and (2S)-flavanones[J]. Metab. Eng., 2016, 38: 47-55. |
| 45 | GALANIE S, THODEY K, TRENCHARD I J, et al. Complete biosynthesis of opioids in yeast[J]. Science, 2015, 349(6252): 1095-1100. |
| 46 | MILKE L, MARIENHAGEN J. Engineering intracellular malonyl-CoA availability in microbial hosts and its impact on polyketide and fatty acid synthesis[J]. Appl. Microbiol. Biotechnol., 2020, 104(14): 6057-6065. |
| 47 | MILKE L, KALLSCHEUER N, KAPPELMANN J, et al. Tailoring Corynebacterium glutamicum towards increased malonyl-CoA availability for efficient synthesis of the plant pentaketide noreugenin[J]. Microb. Cell. Fact., 2019, 18(1): 71. |
| 48 | BRUSSELER C, RADEK A, TENHAEF N, et al. The myo-inositol/proton symporter IolT1 contributes to D-xylose uptake in Corynebacterium glutamicum[J]. Bioresour. Technol., 2018, 249: 953-961. |
| 49 | KALLSCHEUER N, MARIENHAGEN J. Corynebacterium glutamicum as platform for the production of hydroxybenzoic acids[J]. Microbial Cell Factories, 2018, 17(1): 70. |
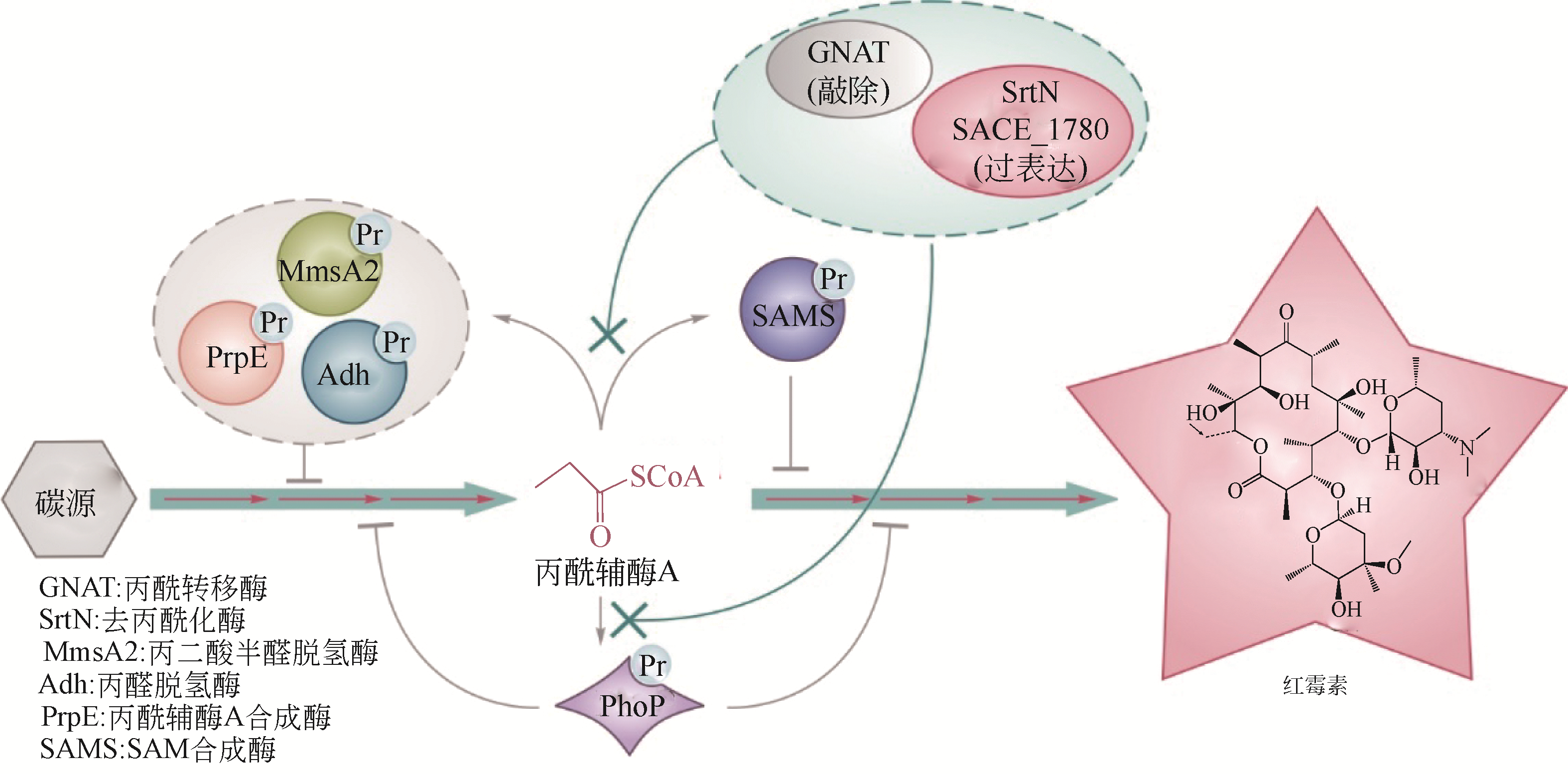
| 50 | XU J Y, XU Y, XU Z, et al. Protein acylation is a general regulatory mechanism in biosynthetic pathway of acyl-CoA-derived natural products[J]. Cell Chemical Biology, 2018, 25(8): 984-995. |
| 51 | YOU D, WANG M M, YIN B C, et al. Precursor supply for erythromycin biosynthesis: engineering of propionate assimilation pathway based on propionylation modification[J]. ACS Synthetic Biology, 2019, 8(2): 371-380. |
| 52 | LIU Q, XIAO L, ZHOU Y, et al. Development of Streptomyces sp. FR-008 as an emerging chassis[J]. Synth. Syst. Biotechnol., 2016, 1(3): 207-214. |
| 53 | BU Q T, YU P, WANG J, et al. Rational construction of genome-reduced and high-efficient industrial Streptomyces chassis based on multiple comparative genomic approaches[J]. Microbial Cell Factories, 2019, 18(1): 1-17. |
| 54 | JIANG W, BIKARD D, COX D, et al. RNA-guided editing of bacterial genomes using CRISPR-Cas systems[J]. Nature Biotechnology, 2013, 31(3): 233-239. |
| 55 | LIU R, DENG Z, LIU T. Streptomyces species: ideal chassis for natural product discovery and overproduction[J]. Metab. Eng., 2018, 50: 74-84. |
| 56 | ZHANG M M, WONG F T, WANG Y, et al. CRISPR–Cas9 strategy for activation of silent Streptomyces biosynthetic gene clusters[J]. Nature Chemical Biology, 2017, 13(6): 607-609. |
| 57 | LIU Y, REN C Y, WEI W P, et al. A CRISPR-Cas9 strategy for activating the Saccharopolyspora erythraea erythromycin biosynthetic gene cluster with knock-in bidirectional promoters[J]. ACS Synth. Biol., 2019, 8(5): 1134-1143. |
| 58 | LIU Y, WEI W P, YE B C. High GC content Cas9-mediated genome-editing and biosynthetic gene cluster activation in Saccharopolyspora erythraea[J]. ACS Synthetic Biology, 2018, 7(5): 1338-1348. |
| 59 | TONG Y J, WHITFORD C M, BLIN K, et al. CRISPR-Cas9, CRISPRi and CRISPR-BEST-mediated genetic manipulation in streptomycetes[J]. Nat. Protoc., 2020, 15(8): 2470-2502. |
| 60 | COBB R E, WANG Y J, ZHAO H M. High-efficiency multiplex genome editing of streptomyces species using an engineered CRISPR/Cas system[J]. ACS Synthetic Biology, 2015, 4(6): 723-728. |
| 61 | LIANG M D, LIU L, WANG W, et al. Simple cloning of large natural product biosynthetic gene cluster by CRISPR/Cas12a-mediated fast direct capturing strategy[J]. BioRxiv, DOI: https//doi.org/10.1101/2020.06.25.170191. |
| 62 | KALLIFIDAS D, JIANG G, DING Y, et al. Rational engineering of Streptomyces albus J1074 for the overexpression of secondary metabolite gene clusters[J]. Microbial Cell Factories, 2018, 17(1): 25. |
| 63 | YOU D, ZHANG B Q, YE B C. GntR family regulator DasR controls acetate assimilation by directly repressing the acsA gene in Saccharopolyspora erythraea[J]. Journal of Bacteriology, 2018, 200(13):e00685. |
| 64 | LIAO C H, XU Y, RIGALI S, et al. DasR is a pleiotropic regulator required for antibiotic production, pigment biosynthesis, and morphological development in Saccharopolyspora erythraea[J]. Appl. Microbiol. Biotechnol., 2015, 99(23): 10215-10224. |
| 65 | LIAO C H, YAO L L, YE B C. Three genes encoding citrate synthases in Saccharopolyspora erythraea are regulated by the global nutrient-sensing regulators GlnR, DasR, and CRP[J]. Molecular Microbiology, 2014, 94(5): 1065-1084. |
| 66 | YOU D, WANG M M, YE B C. Acetyl-CoA synthetases of Saccharopolyspora erythraea are regulated by the nitrogen response regulator GlnR at both transcriptional and post-translational levels[J]. Molecular Microbiology, 2017, 103(5): 845-859. |
| 67 | XU Y, LIAO C H, YAO L L, et al. GlnR and PhoP directly regulate the transcription of genes encoding starch-degrading, amylolytic enzymes in Saccharopolyspora erythraea[J]. Appl. Environ. Microbiol., 2016, 82(23): 6819-6830. |
| 68 | YAO L L, YE B C. Reciprocal regulation of GlnR and PhoP in response to nitrogen and phosphate limitations in Saccharopolyspora erythraea[J]. Appl. Environ. Microbiol., 2016, 82(1): 409-420. |
| 69 | YAO L L, LIAO C H, HUANG G, et al. GlnR-mediated regulation of nitrogen metabolism in the actinomycete Saccharopolyspora erythraea[J]. Appl. Microbiol. Biotechnol., 2014, 98(18): 7935-7948. |
| 70 | LIAO C H, YAO L, XU Y, et al. Nitrogen regulator GlnR controls uptake and utilization of non-phosphotransferase-system carbon sources in actinomycetes[J]. Proceedings of the National Academy of Sciences of the United States of America, 2015, 112(51): 15630-15635. |
| 71 | XU Y, YOU D, YAO L L, et al. Phosphate regulator PhoP directly and indirectly controls transcription of the erythromycin biosynthesis genes in Saccharopolyspora erythraea[J]. Microb. Cell Fact., 2019, 18(1): 206. |
| 72 | MARTIN J F, RODRIGUEZ-GARCIA A, LIRAS P. The master regulator PhoP coordinates phosphate and nitrogen metabolism, respiration, cell differentiation and antibiotic biosynthesis: comparison in Streptomyces coelicolor and Streptomyces avermitilis[J]. Journal of Antibiotics, 2017, 70(5): 534-541. |
| 73 | YANG R, LIU X, WEN Y, et al. The PhoP transcription factor negatively regulates avermectin biosynthesis in Streptomyces avermitilis[J]. Appl. Microbiol. Biotechnol., 2015, 99(24): 10547-10557. |
| 74 | LIU H, MARSAFARI M, WANG F, et al. Engineering acetyl-CoA metabolic shortcut for eco-friendly production of polyketides triacetic acid lactone in Yarrowia lipolytica[J]. Metabolic Engineering, 2019, 56: 60-68. |
| 75 | TANG S Y, QIAN S, AKINTERINWA O, et al. Screening for enhanced triacetic acid lactone production by recombinant Escherichia coli expressing a designed triacetic acid lactone reporter[J]. J. Am .Chem. Soc., 2013, 135(27): 10099-10103. |
| 76 | CARDENAS J, SILVA N A DA. Metabolic engineering of Saccharomyces cerevisiae for the production of triacetic acid lactone[J]. Metab. Eng., 2014, 25: 194-203. |
| 77 | LI Y, QIAN S A, DUNN R, et al. Engineering Escherichia coli to increase triacetic acid lactone (TAL) production using an optimized TAL sensor-reporter system[J]. Journal of Industrial Microbiology & Biotechnology, 2018, 45(9): 789-793. |
| [1] | 李伯耿, 罗英武, 刘平伟. 聚合物产品工程研究内容与方法的思考[J]. 化工进展, 2023, 42(8): 3905-3909. |
| [2] | 孙潇, 朱光涛, 裴爱国. 氢液化装置产业化与研究进展[J]. 化工进展, 2023, 42(3): 1103-1117. |
| [3] | 姚伦, 周雍进. 一碳化合物生物利用和转化研究进展[J]. 化工进展, 2023, 42(1): 16-29. |
| [4] | 郭峰, 张尚杰, 蒋羽佳, 姜万奎, 信丰学, 章文明, 姜岷. 一碳资源在酵母中的利用与转化[J]. 化工进展, 2023, 42(1): 30-39. |
| [5] | 赵路军, 祁雨奇, 邵嘉铭, 褚健, 王智化, 冯毅萍. 流程工业智能制造准备度模型及应用实践[J]. 化工进展, 2023, 42(1): 118-127. |
| [6] | 包淼清. 苯乙烯产品的浙江制造质量标准研究[J]. 化工进展, 2022, 41(S1): 648-655. |
| [7] | 宋望一, 赵新访, 刘伟, 薛玲. 阀门硬化处理工艺对蒽醌法生产双氧水的影响[J]. 化工进展, 2022, 41(S1): 54-59. |
| [8] | 王子宗, 高立兵, 索寒生. 未来石化智能工厂顶层设计: 现状、对比及展望[J]. 化工进展, 2022, 41(7): 3387-3401. |
| [9] | 张凡, 王树众, 李艳辉, 杨健乔, 孙圣瀚. 中国制造业碳排放问题分析与减排对策建议[J]. 化工进展, 2022, 41(3): 1645-1653. |
| [10] | 杨立宁, 郑东昊, 王立新, 杨光. 基于蜻蜓翅脉结构的连续碳纤维增强树脂基复合材料仿生设计与增材制造[J]. 化工进展, 2022, 41(11): 5961-5967. |
| [11] | 汪小钰, 胡平, 操齐高, 李世磊, 胡卜亮, 葛松伟, 杨帆, 陈波, 朱昕宇, 王快社. 不同模式多功能柔性传感器研究进展[J]. 化工进展, 2022, 41(10): 5474-5493. |
| [12] | 陶雨萱, 张尚杰, 景艺文, 信丰学, 董维亮, 周杰, 蒋羽佳, 章文明, 姜岷. 甲基营养型大肠杆菌构建策略的研究进展[J]. 化工进展, 2021, 40(7): 3932-3941. |
| [13] | 郭亮, 高聪, 张丽, 陈修来, 刘立明. 人工代谢路径适配性的研究进展[J]. 化工进展, 2021, 40(3): 1252-1261. |
| [14] | 王颖, 曲俊泽, 梁楠, 郝鹤, 元英进. 合成类胡萝卜素细胞工厂的快速构建和定向进化[J]. 化工进展, 2021, 40(3): 1187-1201. |
| [15] | 孙文涛, 李春. 微生物合成植物天然产物的细胞工厂设计与构建[J]. 化工进展, 2021, 40(3): 1202-1214. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||