化工进展 ›› 2024, Vol. 43 ›› Issue (11): 6356-6371.DOI: 10.16085/j.issn.1000-6613.2023-1981
• 生物与医药化工 • 上一篇
基于PAT技术的动物细胞培养在线检测研究进展
- 北京石油化工学院新材料与化工学院,恩泽生物质精细化工北京市重点实验室,北京 102617
-
收稿日期:2023-11-14修回日期:2024-03-21出版日期:2024-11-15发布日期:2024-12-07 -
通讯作者:周光正,王学重 -
作者简介:周光正(1981—),男,博士,副研究员,研究方向为智能检测与过程模拟。E-mail:zhouguangzheng@bipt.edu.cn。 -
基金资助:北京市自然科学基金(IS23033);国家自然科学基金重点项目(61633006);北京市教育委员会项目(22019821001)
Progress on on-line monitoring for animal cell culture based on PAT technology
ZHOU Guangzheng( ), WANG Xuezhong(
), WANG Xuezhong( )
)
- Beijing Key Laboratory of Enze Biomass Fine Chemicals, College of New Materials and Chemical Engineering, Beijing Institute of Petrochemical Technology, Beijing 102617, China
-
Received:2023-11-14Revised:2024-03-21Online:2024-11-15Published:2024-12-07 -
Contact:ZHOU Guangzheng, WANG Xuezhong
摘要:
动物细胞培养是一项重要的生物工程技术,在该过程中需要对一些重要的生物和生化指标进行监测。目前广泛采用的是取样离线分析手段,存在时间滞后性和潜在的染菌风险。利用过程分析技术(process analytical technology,PAT)在线实时检测动物细胞培养状况,进而实现培养过程的优化与控制,具有重要的学术意义和工业应用价值。本文介绍了两项应用前景最为广阔的细胞培养PAT技术,即原位显微镜成像与拉曼光谱技术,分别用于在线测量细胞培养过程的重要生物指标(细胞数目、尺寸、形状)与生化指标(营养物质、代谢物质、蛋白质产物的浓度)。作为这两项技术成功应用的关键,文中重点介绍了图像分析算法和拉曼光谱建模方法。此外,对近红外光谱、荧光光谱等技术的一些研究探索也做了简要概括性介绍。最后,结合PAT技术的发展,探讨了实现动物细胞培养过程自动化与智能化的未来发展方向。
中图分类号:
引用本文
周光正, 王学重. 基于PAT技术的动物细胞培养在线检测研究进展[J]. 化工进展, 2024, 43(11): 6356-6371.
ZHOU Guangzheng, WANG Xuezhong. Progress on on-line monitoring for animal cell culture based on PAT technology[J]. Chemical Industry and Engineering Progress, 2024, 43(11): 6356-6371.
| 方法 | 代表算法 | 功能 | 优势 | 劣势 |
|---|---|---|---|---|
| 回归算法 | Count-ception[ | 细胞计数 | 数据标注 工作量少 | 精度受图像复杂性 影响较大 |
| 密度估计算法 | FCRN[ | 细胞分布密度图 | 数据标注 工作量适中 | 不提供细胞个体信息 |
| 语义分割算法 | U-Net[ | 像素级别分割 (不区分细胞个体) | 提供细胞分布区域 | 数据标注工作量很大, 不分割细胞聚团 |
| 实例分割算法 | Mask R-CNN[ | 像素级别分割 (区分细胞个体) | 提供细胞个体信息, 包括聚团中个体 | 数据标注工作量最大 |
表1 四种基于深度学习的细胞图像分析方法的比较
| 方法 | 代表算法 | 功能 | 优势 | 劣势 |
|---|---|---|---|---|
| 回归算法 | Count-ception[ | 细胞计数 | 数据标注 工作量少 | 精度受图像复杂性 影响较大 |
| 密度估计算法 | FCRN[ | 细胞分布密度图 | 数据标注 工作量适中 | 不提供细胞个体信息 |
| 语义分割算法 | U-Net[ | 像素级别分割 (不区分细胞个体) | 提供细胞分布区域 | 数据标注工作量很大, 不分割细胞聚团 |
| 实例分割算法 | Mask R-CNN[ | 像素级别分割 (区分细胞个体) | 提供细胞个体信息, 包括聚团中个体 | 数据标注工作量最大 |
| 方法 | 功能 | 数据处理方式 |
|---|---|---|
| 中心化 | 增加光谱间差异 | 减去校正集平均光谱 |
| 平滑 | 提高信噪比 | 基于Savitzky-Golay卷积的 多项式最小二乘拟合 |
| 标准正态变换 | 消除表面散射、 固体颗粒大小等的影响 | 先进行均值中心化, 再除以标准偏差 |
| 求导 | 消除基线和其他 背景的干扰 | 基于Savitzky-Golay卷积的 一阶和二阶导数 |
| 基线校正 | 扣除荧光、黑体辐射等 产生的背景 | 采用自适应迭代重加权惩罚 最小二乘法拟合背景 |
表2 经典的拉曼光谱预处理方法[72-73]
| 方法 | 功能 | 数据处理方式 |
|---|---|---|
| 中心化 | 增加光谱间差异 | 减去校正集平均光谱 |
| 平滑 | 提高信噪比 | 基于Savitzky-Golay卷积的 多项式最小二乘拟合 |
| 标准正态变换 | 消除表面散射、 固体颗粒大小等的影响 | 先进行均值中心化, 再除以标准偏差 |
| 求导 | 消除基线和其他 背景的干扰 | 基于Savitzky-Golay卷积的 一阶和二阶导数 |
| 基线校正 | 扣除荧光、黑体辐射等 产生的背景 | 采用自适应迭代重加权惩罚 最小二乘法拟合背景 |
| 1 | SZKODNY Alana C, LEE Kelvin H. Biopharmaceutical manufacturing: Historical perspectives and future directions[J]. Annual Review of Chemical and Biomolecular Engineering, 2022, 13: 141-165. |
| 2 | Róisín O'FLAHERTY, BERGIN Adam, FLAMPOURI Evangelia, et al. Mammalian cell culture for production of recombinant proteins: A review of the critical steps in their biomanufacturing[J]. Biotechnology Advances, 2020, 43: 107552. |
| 3 | HUNTER Molly, YUAN Ping, VAVILALA Divya, et al. Optimization of protein expression in mammalian cells[J]. Current Protocols in Protein Science, 2019, 95(1): e77. |
| 4 | TRIPATHI Nagesh K, SHRIVASTAVA Ambuj. Recent developments in bioprocessing of recombinant proteins: Expression hosts and process development[J]. Frontiers in Bioengineering and Biotechnology, 2019, 7: 420. |
| 5 | 朱紫瑜, 王冠, 庄英萍. 大规模哺乳动物细胞培养工程的现状与展望[J]. 合成生物学, 2021, 2(4): 612-634. |
| ZHU Ziyu, WANG Guan, ZHUANG Yingping. Present situation and prospect for large-scale mammalian cell culture engineering[J]. Synthetic Biology Journal, 2021, 2(4): 612-634. | |
| 6 | Daniela CADENA-HERRERA, ESPARZA-DE LARA Joshua E, RAMÍREZ-IBAÑEZ Nancy D, et al. Validation of three viable-cell counting methods: Manual, semi-automated, and automated[J]. Biotechnology Reports, 2015, 7: 9-16. |
| 7 | BACUS James W, GRACE Les J. Optical microscope system for standardized cell measurements and analyses[J]. Applied Optics, 1987, 26(16): 3280-3293. |
| 8 | HEIDEMANN Steven R, LAMOUREUX Phillip, Kha NGO, et al. Open-dish incubator for live cell imaging with an inverted microscope[J]. Bio Techniques, 2003, 35(4): 708-714, 716. |
| 9 | ROLINGER Laura, Matthias RÜDT, Jürgen HUBBUCH. A critical review of recent trends, and a future perspective of optical spectroscopy as PAT in biopharmaceutical downstream processing[J]. Analytical and Bioanalytical Chemistry, 2020, 412(9): 2047-2064. |
| 10 | GRANGEIA Helena Bigares, SILVA Cláudia, SIMÕES Sérgio Paulo, et al. Quality by design in pharmaceutical manufacturing: A systematic review of current status, challenges and future perspectives[J]. European Journal of Pharmaceutics and Biopharmaceutics, 2020, 147: 19-37. |
| 11 | RATHORE Anurag S, WINKLE Helen. Quality by design for biopharmaceuticals[J]. Nature Biotechnology, 2009, 27(1): 26-34. |
| 12 | RATHORE Anurag S, KAPOOR Gautam. Application of process analytical technology for downstream purification of biotherapeutics[J]. Journal of Chemical Technology & Biotechnology, 2015, 90(2): 228-236. |
| 13 | SIMON Levente L, PATAKI Hajnalka, MAROSI György, et al. Assessment of recent process analytical technology (PAT) trends: A multiauthor review[J]. Organic Process Research & Development, 2015, 19(1): 3-62. |
| 14 | READ E K, PARK J T, SHAH R B, et al. Process analytical technology (PAT) for biopharmaceutical products: Part Ⅰ. concepts and applications[J]. Biotechnology and Bioengineering, 2010, 105(2): 276-284. |
| 15 | GERZON Gabriella, SHENG Yi, KIRKITADZE Marina. Process Analytical Technologies—Advances in bioprocess integration and future perspectives[J]. Journal of Pharmaceutical and Biomedical Analysis, 2022, 207: 114379. |
| 16 | WASALATHANTHRI Dhanuka P, REHMANN Matthew S, SONG Yuanli, et al. Technology outlook for real-time quality attribute and process parameter monitoring in biopharmaceutical development-a review[J]. Biotechnology and Bioengineering, 2020, 117(10): 3182-3198. |
| 17 | MOWBRAY Max, SAVAGE Thomas, WU Chufan, et al. Machine learning for biochemical engineering: A review[J]. Biochemical Engineering Journal, 2021, 172: 108054. |
| 18 | QI Yaping, HU Dan, JIANG Yucheng, et al. Recent progresses in machine learning assisted Raman spectroscopy[J]. Advanced Optical Materials, 2023, 11(14): 2203104. |
| 19 | HUO Yan, LIU Tao, YANG Yixuan, et al. In situ measurement of 3D crystal size distribution by double-view image analysis with case study on L-glutamic acid crystallization[J]. Industrial & Engineering Chemistry Research, 2020, 59(10): 4646-4658. |
| 20 | WU Yuanyi, GAO Zhenguo, ROHANI Sohrab. Deep learning-based oriented object detection for in situ image monitoring and analysis: A process analytical technology (PAT) application for taurine crystallization[J]. Chemical Engineering Research and Design, 2021, 170: 444-455. |
| 21 | LIU Jian, KUANG Wenjie, LIU Jiaqiang, et al. In-situ multi-phase flow imaging for particle dynamic tracking and characterization: Advances and applications[J]. Chemical Engineering Journal, 2022, 438: 135554. |
| 22 | WANG Xiaoli, ZHOU Guangzheng, LIANG Lipeng, et al. Deep learning-based image analysis for in situ microscopic imaging of cell culture process[J]. Engineering Applications of Artificial Intelligence, 2024, 129: 107621. |
| 23 | REARDON Kenneth F. Practical monitoring technologies for cells and substrates in biomanufacturing[J]. Current Opinion in Biotechnology, 2021, 71: 225-230. |
| 24 | Christian LÜDER, LINDNER Patrick, David BULNES-ABUNDIS, et al. In situ microscopy and MIR-spectroscopy as non-invasive optical sensors for cell cultivation process monitoring[J]. Pharmaceutical Bioprocessing, 2014, 2(2): 157-166. |
| 25 | BITTNER C, WEHNERT G, SCHEPER T. In situ microscopy for on-line determination of biomass[J]. Biotechnology and Bioengineering, 1998, 60(1): 24-35. |
| 26 | ZHAO Liang, FU Hsu-Yuan, ZHOU Weichang, et al. Advances in process monitoring tools for cell culture bioprocesses[J]. Engineering in Life Sciences, 2015, 15(5): 459-468. |
| 27 | MAITRA Mausumi, KUMAR GUPTA Rahul, MUKHERJEE Manali. Detection and counting of red blood cells in blood cell images using Hough transform[J]. International Journal of Computer Applications, 2012, 53(16): 18-22. |
| 28 | ZHOU Xiaogen, LI Zuoyong, XIE Huosheng, et al. Leukocyte image segmentation based on adaptive histogram thresholding and contour detection[J]. Current Bioinformatics, 2020, 15(3): 187-195. |
| 29 | GAMARRA Margarita, ZUREK Eduardo, ESCALANTE Hugo Jair, et al. Split and merge watershed: A two-step method for cell segmentation in fluorescence microscopy images[J]. Biomedical Signal Processing and Control, 2019, 53: 101575. |
| 30 | ZHANG Jiawei, LI Chen, RAHAMAN Md Mamunur, et al. A comprehensive review of image analysis methods for microorganism counting: From classical image processing to deep learning approaches[J]. Artificial Intelligence Review, 2022, 55(4): 2875-2944. |
| 31 | RUDOLPH Guido, LINDNER Patrick, GIERSE Alexander, et al. Online monitoring of microcarrier based fibroblast cultivations with in situ microscopy[J]. Biotechnology and Bioengineering, 2008, 99(1): 136-145. |
| 32 | GUEZ J S, Ph CASSAR J, WARTELLE F, et al. Real time in situ microscopy for animal cell-concentration monitoring during high density culture in bioreactor[J]. Journal of Biotechnology, 2004, 111(3): 335-343. |
| 33 | GUSTAVSSON R, MANDENIUS C F, LÖFGREN S, et al. In situ microscopy as online tool for detecting microbial contaminations in cell culture[J]. Journal of Biotechnology, 2019, 296: 53-60. |
| 34 | VANCLEEF Arne, MAES Dominique, VAN GERVEN Tom, et al. Flow-through microscopy and image analysis for crystallization processes[J]. Chemical Engineering Science, 2022, 248: 117067. |
| 35 | CALDERON DE ANDA J, WANG X Z, ROBERTS K J. Multi-scale segmentation image analysis for the in-process monitoring of particle shape with batch crystallisers[J]. Chemical Engineering Science, 2005, 60(4): 1053-1065. |
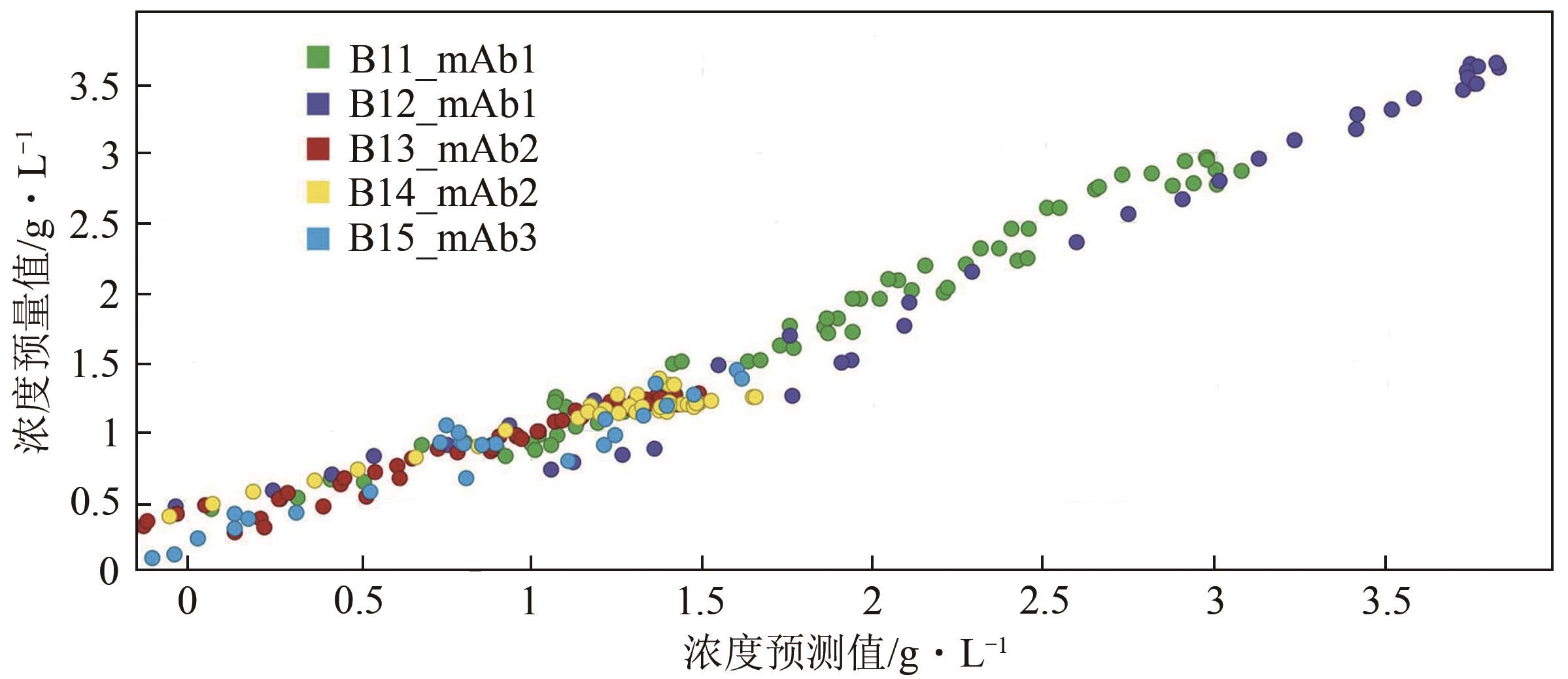
| 36 | Niall O'MAHONY, CAMPBELL Sean, CARVALHO Anderson, et al. Deep learning vs. traditional computer vision[C]//ARAI Kohei, KAPOOR Supriya eds. Advances in Computer Vision: Proceedings of the 2019 Computer Vision Conference (CVC). Cham, Switzerland: Springer Nature Switzerland AG, 2019: 128-144. |
| 37 | LITJENS Geert, KOOI Thijs, BEJNORDI Babak Ehteshami, et al. A survey on deep learning in medical image analysis[J]. Medical Image Analysis, 2017, 42: 60-88. |
| 38 | DODIA Shubham, ANNAPPA B, MAHESH Padukudru A. Recent advancements in deep learning based lung cancer detection: A systematic review[J]. Engineering Applications of Artificial Intelligence, 2022, 116: 105490. |
| 39 | ZHANG Jinghua, LI Chen, YIN Yimin, et al. Applications of artificial neural networks in microorganism image analysis: A comprehensive review from conventional multilayer perceptron to popular convolutional neural network and potential visual transformer[J]. Artificial Intelligence Review, 2023, 56(2): 1013-1070. |
| 40 | NVIDIA. CUDA C++ Programming Guide[EB/OL]. (2023-08-24) [2023-11-08]. . |
| 41 | COHEN Joseph Paul, BOUCHER Genevieve, GLASTONBURY Craig A, et al. Count-ception: Counting by fully convolutional redundant counting[C]//2017 IEEE International Conference on Computer Vision Workshops (ICCVW). October 22-29, 2017, Venice, Italy: IEEE, 2017: 18-26. |
| 42 | STALLMANN Dominik, GÖPFERT Jan P, SCHMITZ Julian, et al. Towards an automatic analysis of CHO-K1 suspension growth in microfluidic single-cell cultivation[J]. Bioinformatics, 2021, 37(20): 3632-3639. |
| 43 | XIE Weidi, Alison NOBLE J, ZISSERMAN Andrew. Microscopy cell counting and detection with fully convolutional regression networks[J]. Computer Methods in Biomechanics and Biomedical Engineering: Imaging & Visualization, 2018, 6(3): 283-292. |
| 44 | HE Shenghua, MINN Kyaw Thu, Lilianna SOLNICA-KREZEL, et al. Deeply-supervised density regression for automatic cell counting in microscopy images[J]. Medical Image Analysis, 2021, 68: 101892. |
| 45 | RONNEBERGER Olaf, FISCHER Philipp, BROX Thomas. U-net: Convolutional networks for biomedical image segmentation[C]// NAVAB Nassir, HORNEGGER Joachim, WELLS William M, et al. International Conference on Medical Image Computing and Computer-Assisted Intervention - MICCAI 2015. Cham: Springer, 2015: 234-241. |
| 46 | ASHA S B, GOPAKUMAR G, SUBRAHMANYAM Gorthi R K Sai. Saliency and ballness driven deep learning framework for cell segmentation in bright field microscopic images[J]. Engineering Applications of Artificial Intelligence, 2023, 118: 105704. |
| 47 | HE Kaiming, GKIOXARI Georgia, Piotr DOLLÁR, et al. Mask R-CNN[C]//2017 IEEE International Conference on Computer Vision (ICCV). October 22-29, 2017, Venice, Italy: IEEE, 2017: 2961-2969. |
| 48 | BOLYA Daniel, ZHOU Chong, XIAO Fanyi, et al. YOLACT++ better real-time instance segmentation[J]. IEEE Transactions on Pattern Analysis and Machine Intelligence, 2022, 44(2): 1108-1121. |
| 49 | FALKO Lavitt, RIJLAARSDAM DEMI J, DER LINDEN DENNET Van, et al. Deep learning and transfer learning for automatic cell counting in microscope images of human cancer cell lines[J]. Applied Sciences, 2021, 11(11): 4912. |
| 50 | 张倩, 王夏黎, 王炜昊, 等. 基于多尺度特征融合的细胞计数方法[J]. 图学学报, 2023, 44(1): 41-49. |
| ZHANG Qian, WANG Xiali, WANG Weihao, et al. Cell counting method based on multi-scale feature fusion[J]. Journal of Graphics, 2023, 44(1): 41-49. | |
| 51 | JIANG Ni, YU Feihong. Multi-column network for cell counting[J]. OSA Continuum, 2020, 3(7): 1834-1846. |
| 52 | GUO Yue, STEIN Jason, WU Guorong, et al. SAU-net: A universal deep network for cell counting[C]//Proceedings of the 10th ACM International Conference on Bioinformatics, Computational Biology and Health Informatics. New York: Association for Computing Machinery, 2019: 299-306. |
| 53 | ASGARI TAGHANAKI Saeid, ABHISHEK Kumar, COHEN Joseph Paul, et al. Deep semantic segmentation of natural and medical images: A review[J]. Artificial Intelligence Review, 2021, 54(1): 137-178. |
| 54 | SHELHAMER Evan, LONG Jonathan, DARRELL Trevor. Fully convolutional networks for semantic segmentation[C]//2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR). Boston: IEEE, 2015: 3431-3440. |
| 55 | FALK Thorsten, Dominic MAI, BENSCH Robert, et al. U-net: Deep learning for cell counting, detection, and morphometry[J]. Nature Methods, 2019, 16(1): 67-70. |
| 56 | HAFIZ Abdul Mueed, BHAT Ghulam Mohiuddin. A survey on instance segmentation: State of the art[J]. International Journal of Multimedia Information Retrieval, 2020, 9(3): 171-189. |
| 57 | REN Shaoqing, HE Kaiming, GIRSHICK Ross, et al. Faster R-CNN: Towards real-time object detection with region proposal networks[J]. IEEE Transactions on Pattern Analysis and Machine Intelligence, 2017, 39(6): 1137-1149. |
| 58 | CAI Zhaowei, VASCONCELOS Nuno. Cascade R-CNN: High quality object detection and instance segmentation[J]. IEEE Transactions on Pattern Analysis and Machine Intelligence, 2021, 43(5): 1483-1498. |
| 59 | GUO Ruohao, NIU Dantong, QU Liao, et al. SOTR: Segmenting objects with transformers[C]//2021 IEEE/CVF International Conference on Computer Vision (ICCV). Montreal, Canada: IEEE, 2021: 7157-7166. |
| 60 | ZHANG Rufeng, TIAN Zhi, SHEN Chunhua, et al. Mask encoding for single shot instance segmentation[C]//2020 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR). Seattle: IEEE, 2020: 10226-10235. |
| 61 | ZONG Shiliang, ZHOU Guangzheng, LI Meng, et al. Deep learning-based on-line image analysis for continuous industrial crystallization processes[J]. Particuology, 2023, 74: 173-183. |
| 62 | I HAQUE Intisar RIZWAN, NEUBERT Jeremiah. Deep learning approaches to biomedical image segmentation[J]. Informatics in Medicine Unlocked, 2020, 18: 100297. |
| 63 | SHORTEN Connor, KHOSHGOFTAAR Taghi M. A survey on image data augmentation for deep learning[J]. Journal of Big Data, 2019, 6(1): 60. |
| 64 | AGGARWAL Alankrita, MITTAL Mamta, BATTINENI Gopi. Generative adversarial network: An overview of theory and applications[J]. International Journal of Information Management Data Insights, 2021, 1(1): 100004. |
| 65 | CRESWELL Antonia, WHITE Tom, DUMOULIN Vincent, et al. Generative adversarial networks: An overview[J]. IEEE Signal Processing Magazine, 2018, 35(1): 53-65. |
| 66 | ZHUANG Fuzhen, QI Zhiyuan, DUAN Keyu, et al. A comprehensive survey on transfer learning[J]. Proceedings of the IEEE, 2021, 109(1): 43-76. |
| 67 | ESMONDE-WHITE Karen A, CUELLAR Maryann, LEWIS Ian R. The role of Raman spectroscopy in biopharmaceuticals from development to manufacturing[J]. Analytical and Bioanalytical Chemistry, 2022, 414(2): 969-991. |
| 68 | PAUDEL Amrit, RAIJADA Dhara, RANTANEN Jukka. Raman spectroscopy in pharmaceutical product design[J]. Advanced Drug Delivery Reviews, 2015, 89: 3-20. |
| 69 | 周昭露, 李杰, 黄生权, 等. 近红外光谱技术在中药质量控制应用中的化学计量学建模: 综述和展望[J]. 化工进展, 2016, 35(6): 1627-1645. |
| ZHOU Zhaolu, LI Jie, HUANG Shengquan, et al. Development of chemometric modelling in the application of NIR to the quality control of Chinese herbal medicine: literature review and future perspectives[J]. Chemical Industry and Engineering Progress, 2016, 35(6): 1627-1645. | |
| 70 | 梁逸曾, 龚范, 俞汝勤. 化学计量学用于中医药研究[J]. 化学进展, 1999, 19(2): 208-212. |
| LIANG Yizeng, GONG Fan, YU Ruqin. Chemometrics applied to researches on Chinese traditional medicine[J]. Progress in Chemistry, 1999, 19(2): 208-212. | |
| 71 | ESMONDE-WHITE Karen A, CUELLAR Maryann, UERPMANN Carsten, et al. Raman spectroscopy as a process analytical technology for pharmaceutical manufacturing and bioprocessing[J]. Analytical and Bioanalytical Chemistry, 2017, 409(3): 637-649. |
| 72 | LASCH Peter. Spectral pre-processing for biomedical vibrational spectroscopy and microspectroscopic imaging[J]. Chemometrics and Intelligent Laboratory Systems, 2012, 117: 100-114. |
| 73 | GAUTAM Rekha, VANGA Sandeep, ARIESE Freek, et al. Review of multidimensional data processing approaches for Raman and infrared spectroscopy[J]. EPJ Techniques and Instrumentation, 2015, 2(1): 8. |
| 74 | 褚小立. 现代光谱分析中的化学计量学方法[M]. 北京: 化学工业出版社, 2022. |
| CHU Xiaoli. Chemometric methods in modern spectral analysis[M]. Beijing: Chemical Industry Press, 2022. | |
| 75 | HAENLEIN Michael, KAPLAN Andreas M. A beginner's guide to partial least squares analysis[J]. Understanding Statistics, 2004, 3(4): 283-297. |
| 76 | Åsmund RINNAN. Pre-processing in vibrational spectroscopy-when, why and how[J]. Analytical Methods, 2014, 6(18): 7124-7129. |
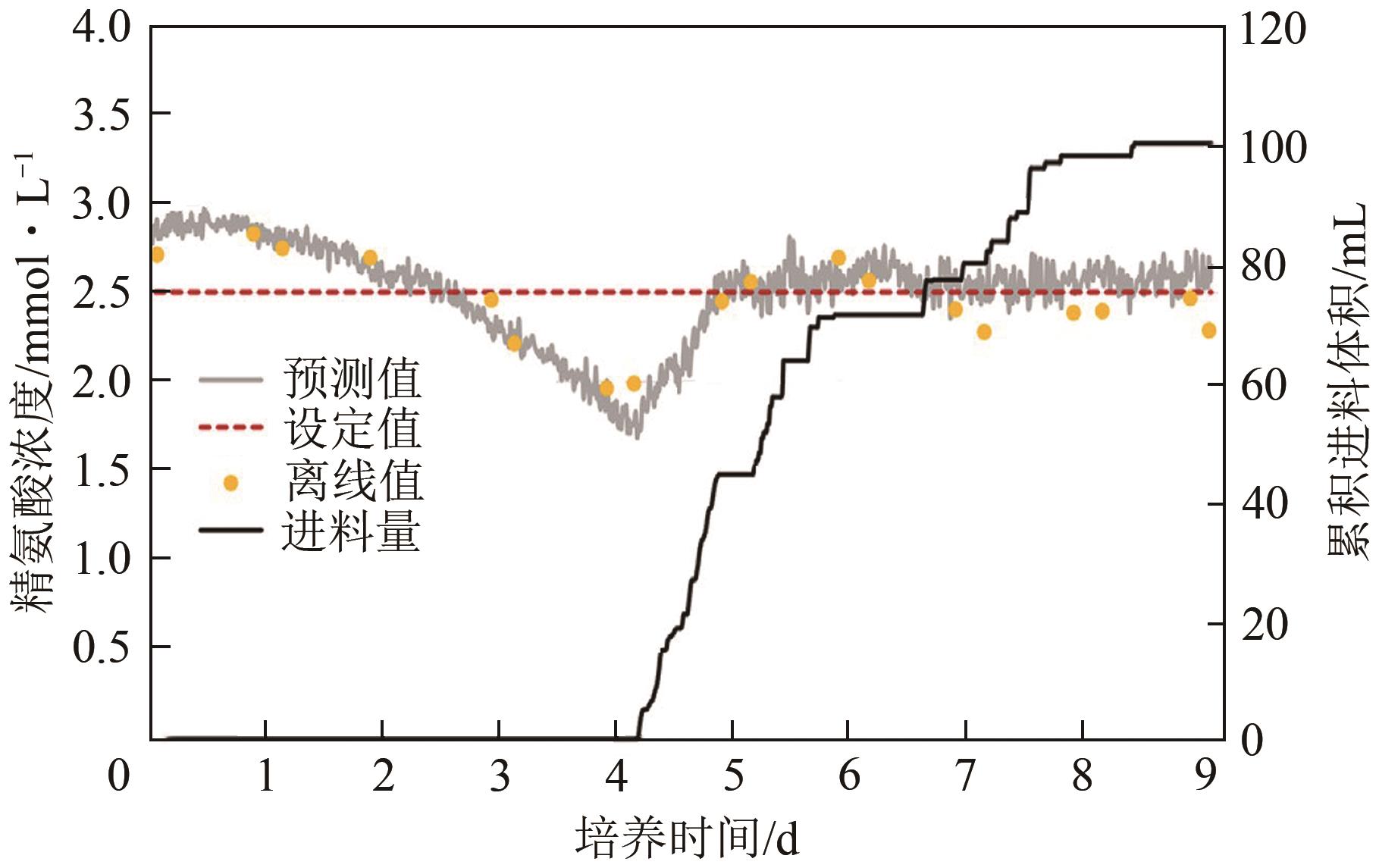
| 77 | MATTHEWS Thomas E, BERRY Brandon N, SMELKO John, et al. Closed loop control of lactate concentration in mammalian cell culture by Raman spectroscopy leads to improved cell density, viability, and biopharmaceutical protein production[J]. Biotechnology and Bioengineering, 2016, 113(11): 2416-2424. |
| 78 | 阎续, 沈丽娟, 胥文彦, 等. 拉曼光谱用于CHO细胞培养液多指标快速分析[J]. 高校化学工程学报, 2019, 33(4): 872-877. |
| YAN Xu, SHEN Lijuan, XU Wenyan, et al. Rapid analysis of multiple parameters of CHO cell culture media using Raman spectroscopy[J]. Journal of Chemical Engineering of Chinese Universities, 2019, 33(4): 872-877. | |
| 79 | BHATIA Hemlata, MEHDIZADEH Hamidreza, DRAPEAU Denis, et al. In-line monitoring of amino acids in mammalian cell cultures using Raman spectroscopy and multivariate chemometrics models[J]. Engineering in Life Sciences, 2018, 18(1): 55-61. |
| 80 | RAFFERTY Carl, JOHNSON Kjell, Jim O'MAHONY, et al. Analysis of chemometric models applied to Raman spectroscopy for monitoring key metabolites of cell culture[J]. Biotechnology Progress, 2020, 36(4): e2977. |
| 81 | MATTHEWS Thomas E, SMELKO John P, BERRY Brandon, et al. Glucose monitoring and adaptive feeding of mammalian cell culture in the presence of strong autofluorescence by near infrared Raman spectroscopy[J]. Biotechnology Progress, 2018, 34(6): 1574-1580. |
| 82 | KAZEMZADEH Mohammadrahim, Miguel MARTINEZ-CALDERON, XU Weiliang, et al. Cascaded deep convolutional neural networks as improved methods of preprocessing Raman spectroscopy data[J]. Analytical Chemistry, 2022, 94(37): 12907-12918. |
| 83 | ABU-ABSI Nicholas R, KENTY Brian M, CUELLAR Maryann Ehly, et al. Real time monitoring of multiple parameters in mammalian cell culture bioreactors using an in-line Raman spectroscopy probe[J]. Biotechnology and Bioengineering, 2011, 108(5): 1215-1221. |
| 84 | Júlia DOMJÁN, PANTEA Eszter, Martin GYÜRKÉS, et al. Real-time amino acid and glucose monitoring system for the automatic control of nutrient feeding in CHO cell culture using Raman spectroscopy[J]. Biotechnology Journal, 2022, 17(5): e2100395. |
| 85 | TULSYAN Aditya, SCHORNER Gregg, KHODABANDEHLOU Hamid, et al. A machine-learning approach to calibrate generic Raman models for real-time monitoring of cell culture processes[J]. Biotechnology and Bioengineering, 2019, 116(10): 2575-2586. |
| 86 | CHEN Gong, HU Jun, QIN Yongjun, et al. Viable cell density on-line auto-control in perfusion cell culture aided by in-situ Raman spectroscopy[J]. Biochemical Engineering Journal, 2021, 172: 108063. |
| 87 | Silvère ANDRÉ, SAINT CRISTAU Lydia, GAILLARD Sabine, et al. In-line and real-time prediction of recombinant antibody titer by in situ Raman spectroscopy[J]. Analytica Chimica Acta, 2015, 892: 148-152. |
| 88 | LI Mengyao, EBEL Bruno, PARIS Cédric, et al. Real-time monitoring of antibody glycosylation site occupancy by in situ Raman spectroscopy during bioreactor CHO cell cultures[J]. Biotechnology Progress, 2018, 34(2): 486-493. |
| 89 | RAFFERTY Carl, Jim O'MAHONY, BURGOYNE Barbara, et al. Raman spectroscopy as a method to replace off-line pH during mammalian cell culture processes[J]. Biotechnology and Bioengineering, 2020, 117(1): 146-156. |
| 90 | WHELAN Jessica, CRAVEN Stephen, GLENNON Brian. In situ Raman spectroscopy for simultaneous monitoring of multiple process parameters in mammalian cell culture bioreactors[J]. Biotechnology Progress, 2012, 28(5): 1355-1362. |
| 91 | BERRY Brandon, MORETTO Justin, MATTHEWS Thomas, et al. Cross-scale predictive modeling of CHO cell culture growth and metabolites using Raman spectroscopy and multivariate analysis[J]. Biotechnology Progress, 2015, 31(2): 566-577. |
| 92 | MEHDIZADEH Hamidreza, LAURI David, KARRY Krizia M, et al. Generic Raman-based calibration models enabling real-time monitoring of cell culture bioreactors[J]. Biotechnology Progress, 2015, 31(4): 1004-1013. |
| 93 | YILMAZ Denizhan, MEHDIZADEH Hamidreza, NAVARRO Dunie, et al. Application of Raman spectroscopy in monoclonal antibody producing continuous systems for downstream process intensification[J]. Biotechnology Progress, 2020, 36(3): e2947. |
| 94 | SCHWARZ Hubert, MÄKINEN Meeri E, CASTAN Andreas, et al. Monitoring of amino acids and antibody N-glycosylation in high cell density perfusion culture based on Raman spectroscopy[J]. Biochemical Engineering Journal, 2022, 182: 108426. |
| 95 | CHONG Il-Gyo, Chi-Hyuck JUN. Performance of some variable selection methods when multicollinearity is present[J]. Chemometrics and Intelligent Laboratory Systems, 2005, 78(1/2): 103-112. |
| 96 | MEHMOOD Tahir, LILAND Kristian Hovde, SNIPEN Lars, et al. A review of variable selection methods in partial least squares regression[J]. Chemometrics and Intelligent Laboratory Systems, 2012, 118: 62-69. |
| 97 | KATOCH Sourabh, CHAUHAN Sumit Singh, KUMAR Vijay. A review on genetic algorithm: Past, present, and future[J]. Multimedia Tools and Applications, 2021, 80(5): 8091-8126. |
| 98 | BOCKLITZ Thomas, WALTER Angela, HARTMANN Katharina, et al. How to pre-process Raman spectra for reliable and stable models?[J]. Analytica Chimica Acta, 2011, 704(1/2): 47-56. |
| 99 | DORIGO Marco, Thomas STÜTZLE. Ant colony optimization: Overview and recent advances[M]//Gendreau Michel, Potvin Jean-Yves. Third Edition. Handbook of Metaheuristics. Cham: Springer, 2019: 311-351. |
| 100 | LI Boyan, Bryan H RAY, LEISTER Kirk J, et al. Performance monitoring of a mammalian cell based bioprocess using Raman spectroscopy[J]. Analytica Chimica Acta, 2013, 796: 84-91. |
| 101 | TULSYAN Aditya, WANG Tony, SCHORNER Gregg, et al. Automatic real-time calibration, assessment, and maintenance of generic Raman models for online monitoring of cell culture processes[J]. Biotechnology and Bioengineering, 2020, 117(2): 406-416. |
| 102 | KOZMA Bence, András SALGÓ, GERGELY Szilveszter. Comparison of multivariate data analysis techniques to improve glucose concentration prediction in mammalian cell cultivations by Raman spectroscopy[J]. Journal of Pharmaceutical and Biomedical Analysis, 2018, 158: 269-279. |
| 103 | ZAVALA-ORTIZ Daniel A, DENNER Aurélia, AGUILAR-USCANGA Maria G, et al. Comparison of partial least square, artificial neural network, and support vector regressions for real-time monitoring of CHO cell culture processes using in situ near-infrared spectroscopy[J]. Biotechnology and Bioengineering, 2022, 119(2): 535-549. |
| 104 | LI Dengshan, LI Lina. Detection of water pH using visible near-infrared spectroscopy and one-dimensional convolutional neural network[J]. Sensors, 2022, 22(15): 5809. |
| 105 | MA Danying, SHANG Linwei, TANG Jinlan, et al. Classifying breast cancer tissue by Raman spectroscopy with one-dimensional convolutional neural network[J]. Spectrochimica Acta Part A: Molecular and Biomolecular Spectroscopy, 2021, 256: 119732. |
| 106 | 赵勇, 何梦园, 王泊林, 等. 基于一维卷积神经网络和拉曼光谱的肺炎支原体菌株分类[J]. 光谱学与光谱分析, 2022, 42(5): 1439-1444. |
| ZHAO Yong, HE Mengyuan, WANG Bolin, et al. Classification of mycoplasma pneumoniae strains based on one-dimensional convolutional neural network and Raman spectroscopy[J]. Spectroscopy and Spectral Analysis, 2022, 42(5): 1439-1444. | |
| 107 | GOODFELLOW Ian, Jean POUGET-ABADIE, MIRZA Mehdi, et al. Generative adversarial networks[J]. Communications of the ACM, 2020, 63(11): 139-144. |
| 108 | GUI Jie, SUN Zhenan, WEN Yonggang, et al. A review on generative adversarial networks: Algorithms, theory, and applications[J]. IEEE Transactions on Knowledge and Data Engineering, 2023, 35(4): 3313-3332. |
| 109 | YU Shixiang, LI Hanfei, LI Xin, et al. Classification of pathogens by Raman spectroscopy combined with generative adversarial networks[J]. Science of the Total Environment, 2020, 726: 138477. |
| 110 | DU Yuwan, HAN Dianpeng, LIU Sha, et al. Raman spectroscopy-based adversarial network combined with SVM for detection of foodborne pathogenic bacteria[J]. Talanta, 2022, 237: 122901. |
| 111 | WU Man, WANG Shuwen, PAN Shirui, et al. Deep learning data augmentation for Raman spectroscopy cancer tissue classification[J]. Scientific Reports, 2021, 11(1): 23842. |
| 112 | MIN Rui, WANG Zhi, ZHUANG Yingping, et al. Application of semi-supervised convolutional neural network regression model based on data augmentation and process spectral labeling in Raman predictive modeling of cell culture processes[J]. Biochemical Engineering Journal, 2023, 191: 108774. |
| 113 | 王天云, 张俊河, 林艳, 等. 动物细胞培养及培养基制备[M]. 北京: 化学工业出版社, 2022. |
| WANG Tianyun, ZHANG Junhe, LIN Yan, et al. Animal cell culture and culture medium preparation[M]. Beijing: Chemical Industry Press, 2022. | |
| 114 | GRAF A, LEMKE J, SCHULZE M, et al. A novel approach for non-invasive continuous in-line control of perfusion cell cultivations by Raman spectroscopy[J]. Frontiers in Bioengineering and Biotechnology, 2022, 10: 719614. |
| 115 | SOMMEREGGER Wolfgang, SISSOLAK Bernhard, KANDRA Kulwant, et al. Quality by control: towards model predictive control of mammalian cell culture bioprocesses[J]. Biotechnology Journal, 2017, 12(7): 1600546. |
| 116 | LUO Yu, KURIAN Varghese, OGUNNAIKE Babatunde A. Bioprocess systems analysis, modeling, estimation, and control[J]. Current Opinion in Chemical Engineering, 2021, 33: 100705. |
| 117 | RAMKRISHNA Doraiswami, SINGH Meenesh R. Population balance modeling: Current status and future prospects[J]. Annual Review of Chemical and Biomolecular Engineering, 2014, 5: 123-146. |
| 118 | MA Yiming, WU Songgu, MACARINGUE Estevao Genito Joao, et al. Recent progress in continuous crystallization of pharmaceutical products: Precise preparation and control[J]. Organic Process Research & Development, 2020, 24(10): 1785-1801. |
| 119 | YANG Li, ZHANG Yan, LIU Peng, et al. Kinetics and population balance modeling of antisolvent crystallization of polymorphic indomethacin[J]. Chemical Engineering Journal, 2022, 428: 132591. |
| 120 | ALHUTHALI Sakhr, KONTORAVDI Cleo. Population balance modelling captures host cell protein dynamics in CHO cell cultures[J]. PLoS One, 2022, 17(3): e0265886. |
| 121 | KARRA Srinivas, SAGER Brian, Nazmul KARIM M. Multi-scale modeling of heterogeneities in mammalian cell culture processes[J]. Industrial & Engineering Chemistry Research, 2010, 49(17): 7990-8006. |
| 122 | SIDOLI Fabio R, ASPREY Steven P, MANTALARIS Athanasios. A coupled single cell-population-balance model for mammalian cell cultures[J]. Industrial & Engineering Chemistry Research, 2006, 45(16): 5801-5811. |
| 123 | RASHEDI Mohammad, RAFIEI Mina, DEMERS Matthew, et al. Machine learning-based model predictive controller design for cell culture processes[J]. Biotechnology and Bioengineering, 2023, 120(8): 2144-2159. |
| 124 | Alison ARNOLD S, CROWLEY John, WOODS Nigel, et al. In-situ near infrared spectroscopy to monitor key analytes in mammalian cell cultivation[J]. Biotechnology and Bioengineering, 2003, 84(1): 13-19. |
| 125 | CLAVAUD Matthieu, ROGGO Yves, VON DAENIKEN Ralph, et al. Chemometrics and in-line near infrared spectroscopic monitoring of a biopharmaceutical Chinese hamster ovary cell culture: Prediction of multiple cultivation variables[J]. Talanta, 2013, 111: 28-38. |
| 126 | QIU Jiang, ARNOLD Mark A, MURHAMMER David W. On-line near infrared bioreactor monitoring of cell density and concentrations of glucose and lactate during insect cell cultivation[J]. Journal of Biotechnology, 2014, 173: 106-111. |
| 127 | KOZMA Bence, András SALGÓ, GERGELY Szilveszter. On-line glucose monitoring by near infrared spectroscopy during the scale up steps of mammalian cell cultivation process development[J]. Bioprocess and Biosystems Engineering, 2019, 42(6): 921-932. |
| 128 | ZAVALA-ORTIZ Daniel A, EBEL Bruno, LI Mengyao, et al. Interest of locally weighted regression to overcome nonlinear effects during in situ NIR monitoring of CHO cell culture parameters and antibody glycosylation[J]. Biotechnology Progress, 2020, 36(1): e2924. |
| 129 | ZAVALA-ORTIZ Daniel Arturo, EBEL Bruno, LI Mengyao, et al. Support Vector and Locally Weighted regressions to monitor monoclonal antibody glycosylation during CHO cell culture processes, an enhanced alternative to Partial Least Squares regression[J]. Biochemical Engineering Journal, 2020, 154: 107457. |
| 130 | KAMBAYASHI Takuya, NOGUCHI Toshimitsu, NOJIMA Akihiro, et al. Glucose monitoring in cell culture with online ultrasound-assisted near-infrared spectroscopy[J]. Analytical Chemistry, 2020, 92(4): 2946-2952. |
| 131 | RHIEL Martin, DUCOMMUN Paul, BOLZONELLA Ivan, et al. Real-time in situ monitoring of freely suspended and immobilized cell cultures based on mid-infrared spectroscopic measurements[J]. Biotechnology and Bioengineering, 2002, 77(2): 174-185. |
| 132 | CAPITO Florian, ZIMMER Aline, SKUDAS Romas. Mid-infrared spectroscopy-based analysis of mammalian cell culture parameters[J]. Biotechnology Progress, 2015, 31(2): 578-584. |
| 133 | TEIXEIRA Ana P, PORTUGAL Carla A M, CARINHAS Nuno, et al. In situ 2D fluorometry and chemometric monitoring of mammalian cell cultures[J]. Biotechnology and Bioengineering, 2009, 102(4): 1098-1106. |
| 134 | TEIXEIRA Ana P, DUARTE Tiago M, CARRONDO M J T, et al. Synchronous fluorescence spectroscopy as a novel tool to enable PAT applications in bioprocesses[J]. Biotechnology and Bioengineering, 2011, 108(8): 1852-1861. |
| 135 | Jens CLAßEN, GRAF Alexander, AUPERT Florian, et al. A novel LED-based 2D-fluorescence spectroscopy system for in-line bioprocess monitoring of Chinese hamster ovary cell cultivations—part Ⅱ[J]. Engineering in Life Sciences, 2019, 19(5): 341-351. |
| 136 | TAKAHASHI Maria Beatriz, LEME Jaci, CARICATI Celso Pereira, et al. Artificial neural network associated to UV/Vis spectroscopy for monitoring bioreactions in biopharmaceutical processes[J]. Bioprocess and Biosystems Engineering, 2015, 38(6): 1045-1054. |
| 137 | DRIESCHNER Tobias, OSTERTAG Edwin, BOLDRINI Barbara, et al. Direct optical detection of cell density and viability of mammalian cells by means of UV/Vis spectroscopy[J]. Analytical and Bioanalytical Chemistry, 2020, 412(14): 3359-3371. |
| 138 | MOORE Brandon, SANFORD Ryan, ZHANG An. Case study: The characterization and implementation of dielectric spectroscopy (biocapacitance) for process control in a commercial GMP CHO manufacturing process[J]. Biotechnology Progress, 2019, 35(3): e2782. |
| 139 | METZE Sabrina, BLIOCH Stefanie, MATUSZCZYK Jens, et al. Multivariate data analysis of capacitance frequency scanning for online monitoring of viable cell concentrations in small-scale bioreactors[J]. Analytical and Bioanalytical Chemistry, 2020, 412(9): 2089-2102. |
| 140 | PARK Seo-Young, PARK Cheol-Hwan, CHOI Dong-Hyuk, et al. Bioprocess digital twins of mammalian cell culture for advanced biomanufacturing[J]. Current Opinion in Chemical Engineering, 2021, 33: 100702. |
| 141 | CHOPDA Viki, GYORGYPAL Aron, YANG Ou, et al. Recent advances in integrated process analytical techniques, modeling, and control strategies to enable continuous biomanufacturing of monoclonal antibodies[J]. Journal of Chemical Technology & Biotechnology, 2022, 97(9): 2317-2335. |
| [1] | 戴征舒, 左元浩, 陈孝罗, 张犁, 赵根, 张学军, 张华. 机器学习在喷射器研究中的应用进展[J]. 化工进展, 2024, 43(S1): 1-12. |
| [2] | 毛宁轩, 万小维, 鞠杰, 胡彦杰, 江浩. 工业气固流化床内流场的CFD-PBM数值模拟和结构优化[J]. 化工进展, 2024, 43(S1): 13-20. |
| [3] | 杨俊辉, 袁君, 张继达, 王金海, 乔红斌, 蔡振义, 马中成. 新型蓄热体结构设计及性能分析[J]. 化工进展, 2024, 43(S1): 282-294. |
| [4] | 周渝, 夏太阳, 韦奇, 唐甜, 田磊. 微通道耦合反渗透膜串联处理甲醇制烯烃废水工艺优化[J]. 化工进展, 2024, 43(S1): 43-51. |
| [5] | 陈王觅, 席北斗, 李鸣晓, 叶美瀛, 侯佳奇, 于承泽, 魏域芳, 孟繁华. 热解系统碳排放削减技术研究进展[J]. 化工进展, 2024, 43(S1): 479-503. |
| [6] | 柳宏伟, 董国亮, 温彦博, 王强华, 徐庆, 王兴盛, 李旭升, 巩洁平, 赵斌, 刘梦瑶. 华亭挤压造粒机组工艺优化[J]. 化工进展, 2024, 43(S1): 52-60. |
| [7] | 陶一, 张晨, 胡益炯, 邱彤. 基于分子结构分布的减压蜡油分子重构模型[J]. 化工进展, 2024, 43(S1): 71-76. |
| [8] | 罗世发, 王侃, 张冰剑, 陈清林. 原油常减压蒸馏装置热集成方案分析评价[J]. 化工进展, 2024, 43(9): 4810-4816. |
| [9] | 孙启超, 聂美华, 伍联营, 胡仰栋. 风光储一体化水电联产系统的优化设计及调度[J]. 化工进展, 2024, 43(9): 4882-4891. |
| [10] | 王禹程, 郭雄, 栾昕奇, 周健, 李想, 幸林广, 周雪芸, 刘颖, 汪德勇, 吴雪娟, 潘琦, 刘建新, 赵祯霞, 赵钟兴. 维生素U生产工艺优化及其热分解机理[J]. 化工进展, 2024, 43(9): 5157-5167. |
| [11] | 王亚男, 刘琳琳, 庄钰, 都健. 基于代理模型的环氧乙烷制乙二醇工艺优化同步热集成[J]. 化工进展, 2024, 43(9): 5234-5241. |
| [12] | 赵星程, 贾方旭, 刘晨雨, 韩宝红, 梅宁, 姚宏. 实际规模PN/A工艺载体挂膜性能与微生物群落[J]. 化工进展, 2024, 43(9): 5242-5249. |
| [13] | 吴宇琦, 李江涛, 丁建智, 宋秀兰, 苏冰琴. 焙烧镁铝水滑石脱除厌氧消化沼气中CO2的效果及机制[J]. 化工进展, 2024, 43(9): 5250-5261. |
| [14] | 何海霞, 万亚萌, 李帆帆, 牛心雨, 张静雯, 李涛, 任保增. 盐酸萘甲唑啉在甲醇-乙酸乙酯体系中的动力学及结晶工艺[J]. 化工进展, 2024, 43(8): 4230-4245. |
| [15] | 张笑恬, 刘思琪, 崔国民, 黄晓璜, 段欢欢, 王金阳. 基于定向协调策略改进换热单元优化的换热网络综合[J]. 化工进展, 2024, 43(8): 4342-4353. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||