化工进展 ›› 2022, Vol. 41 ›› Issue (12): 6531-6539.DOI: 10.16085/j.issn.1000-6613.2022-0459
重组大肠杆菌1,2,4-丁三醇合成途径的平衡优化
- 江南大学工业生物技术教育部重点实验室,生物工程学院,工业微生物研究中心,江苏 无锡 214122
-
收稿日期:2022-03-23修回日期:2022-04-12出版日期:2022-12-20发布日期:2022-12-29 -
通讯作者:诸葛斌 -
作者简介:郭超(1996—),男,硕士研究生,研究方向为微生物代谢工程。E-mail:13008078449@163.com。
Balanced optimization of the 1,2,4-butanetriol synthesis pathway in recombinant Escherichia coli
GUO Chao( ), FENG Ao, LU Xinyao, ZONG Hong, ZHUGE Bin(
), FENG Ao, LU Xinyao, ZONG Hong, ZHUGE Bin( )
)
- Key Laboratory of Industrial Biotechnology, Ministry of Education, Jiangnan University, Laboratory of Industrial Microorganisms, School of Biotechnology, Jiangnan University, Wuxi 214122, Jiangsu, China
-
Received:2022-03-23Revised:2022-04-12Online:2022-12-20Published:2022-12-29 -
Contact:ZHUGE Bin
摘要:
1,2,4-丁三醇(1,2,4-butantriol,BT)属于非天然化学品,是军工材料1,2,4-丁三醇三硝酸酯的前体。在重组大肠杆菌中,木糖经脱氢、脱水、脱羧和还原合成BT。但途径各反应不平衡使得中间代谢物积累限制菌体生长和产物合成。本研究首先通过CRISPR/Cas9敲除基因yjhG和yqhD构建无本底表达宿主菌,随后利用不同启动子组合调节BT合成途径中基因xdh、yjhG和yqhD的表达。结果发现,以P inv 表达醇脱氢酶基因yqhD使BT产量达到14.4g/L;以P tac 表达脱氢酶基因xdh和P rrnH P1表达脱水酶基因yjhG的组合方式,BT产量达到15.6g/L,比对照菌株KXW3009分别增加5.9%和14.7%。本研究通过对中间代谢物木糖酸合成和代谢的组合优化,促进了BT的合成,为后续研究提供了借鉴。
中图分类号:
引用本文
郭超, 冯奥, 陆信曜, 宗红, 诸葛斌. 重组大肠杆菌1,2,4-丁三醇合成途径的平衡优化[J]. 化工进展, 2022, 41(12): 6531-6539.
GUO Chao, FENG Ao, LU Xinyao, ZONG Hong, ZHUGE Bin. Balanced optimization of the 1,2,4-butanetriol synthesis pathway in recombinant Escherichia coli[J]. Chemical Industry and Engineering Progress, 2022, 41(12): 6531-6539.
| 菌株和质粒 | 相关特性 | 来源 |
|---|---|---|
| E.coli W3009(W3009) | E.coli W3110 ΔxylABΔyagEΔyjhHΔptsHIΔmgsA | 实验室保存 |
| E.coli W3010(W3010) | E.coli W3009 ΔyjhGΔyqhD | 本研究 |
| E.coli KXW3009(KXW3009) | E.coli W3009,pEtac-kivD-tac-xdh | 实验室保存 |
| E.coli KXW3010(KXW3010) | E.coli W3010,pEtac-kivD-tac-xdh | 本研究 |
| E.coli KXW3010P0(P0) | E.coli KXW3010,pRSFDuet-P yjhG -yjhG | 本研究 |
| E.coli KXW3010P1(P1) | E.coli KXW3010,pRSFDuet-P yjhG -yjhG-P yqhD-yqhD | 本研究 |
| E.coli KXW3010P2(P2) | E.coli KXW3010,pRSFDuet-P yjhG -yjhG-tac-yqhD | 本研究 |
| E.coli KXW3010P3(P3) | E.coli KXW3010,pRSFDuet-P yjhG -yjhG-inv-yqhD | 本研究 |
| E.coli KXW3010P4(P4) | E.coli KXW3010,pRSFDuet-P yjhG -yjhG-dnaKJ-yqhD | 本研究 |
| E.coli KXW3010P5(P5) | E.coli KXW3010,pRSFDuet-P yjhG -yjhG-grpE-yqhD | 本研究 |
| E.coli KXW3010P3-12(P12) | E.coli W3010,pRSF-P yjhG -yjhG-inv-yqhD,pEtac-kivD-rpsA P1-xdh | 本研究 |
| E.coli KXW3010P3-13(P13) | E.coli W3010,pRSF-P yjhG -yjhG-inv-yqhD,pEtac-kivD-rpsT P1-xdh | 本研究 |
| E.coli KXW3010P3-14(P14) | E.coli W3010,pRSF-P yjhG -yjhG-inv-yqhD,pEtac-kivD-inv-xdh | 本研究 |
| E.coli KXW3010P3-15(P15) | E.coli W3010,pRSF-P yjhG -yjhG-inv-yqhD,pEtac-kivD-dnaKJ-xdh | 本研究 |
| E.coli KXW3010P3-16(P16) | E.coli W3010,pRSF-P yjhG -yjhG-inv-yqhD,pEtac-kivD-grpE-xdh | 本研究 |
| E.coli KXW3010P3-21(P21) | E.coli W3010,pRSFtac-yjhG-inv-yqhD,pEtac-kivD-tac-xdh | 本研究 |
| E.coli KXW3010P3-22(P22) | E.coli W3010,pRSFtac-yjhG-inv-yqhD,pEtac-kivD-rpsA P1-xdh | 本研究 |
| E.coli KXW3010P3-23(P23) | E.coli W3010,pRSFtac-yjhG-inv-yqhD,pEtac-kivD-rpsT P1-xdh | 本研究 |
| E.coli KXW3010P3-24(P24) | E.coli W3010,pRSFtac-yjhG-inv-yqhD,pEtac-kivD-inv-xdh | 本研究 |
| E.coli KXW3010P3-25(P25) | E.coli W3010,pRSFtac-yjhG-inv-yqhD,pEtac-kivD-dnaKJ-xdh | 本研究 |
| E.coli KXW3010P3-26(P26) | E.coli W3010,pRSFtac-yjhG-inv-yqhD,pEtac-kivD-grpE-xdh | 本研究 |
| E.coli KXW3010P3-31(P31) | E.coli W3010,pRSFrrnC P1-yjhG-inv-yqhD,pEtac-kivD-tac-xdh | 本研究 |
| E.coli KXW3010P3-32(P32) | E.coli W3010,pRSFrrnC P1-yjhG-inv-yqhD,pEtac-kivD-rpsA P1-xdh | 本研究 |
| E.coli KXW3010P3-33(P33) | E.coli W3010,pRSFrrnC P1-yjhG-inv-yqhD,pEtac-kivD-rpsT P1-xdh | 本研究 |
| E.coli KXW3010P3-34(P34) | E.coli W3010,pRSFrrnC P1-yjhG-inv-yqhD,pEtac-kivD-inv-xdh | 本研究 |
| E.coli KXW3010P3-35(P35) | E.coli W3010,pRSFrrnC P1-yjhG-inv-yqhD,pEtac-kivD-dnaKJ-xdh | 本研究 |
| E.coli KXW3010P3-36(P36) | E.coli W3010,pRSFrrnC P1-yjhG-inv-yqhD,pEtac-kivD-grpE-xdh | 本研究 |
| E.coli KXW3010P3-41(P41) | E.coli W3010,pRSFrrnH P1-yjhG-inv-yqhD,pEtac-kivD-tac-xdh | 本研究 |
| E.coli KXW3010P3-42(P42) | E.coli W3010,pRSFrrnH P1-yjhG-inv-yqhD,pEtac-kivD-rpsA P1-xdh | 本研究 |
| E.coli KXW3010P3-43(P43) | E.coli W3010,pRSFrrnH P1-yjhG-inv-yqhD,pEtac-kivD-rpsT P1-xdh | 本研究 |
| E.coli KXW3010P3-44(P44) | E.coli W3010,pRSFrrnH P1-yjhG-inv-yqhD,pEtac-kivD-inv-xdh | 本研究 |
| E.coli KXW3010P3-45(P45) | E.coli W3010,pRSFrrnH P1-yjhG-inv-yqhD,pEtac-kivD-dnaKJ-xdh | 本研究 |
| E.coli KXW3010P3-46(P46) | E.coli W3010,pRSFrrnH P1-yjhG-inv-yqhD,pEtac-kivD-grpE-xdh | 本研究 |
| E.coli KXW3010P3-51(P51) | E.coli W3010,pRSFdnaKJrpsT -yjhG-inv-yqhD,pEtac-kivD-tac-xdh | 本研究 |
| E.coli KXW3010P3-52(P52) | E.coli W3010,pRSFdnaKJrpsT -yjhG-inv-yqhD,pEtac-kivD-rpsA P1-xdh | 本研究 |
| E.coli KXW3010P3-53(P53) | E.coli W3010,pRSFdnaKJrpsT -yjhG-inv-yqhD,pEtac-kivD-rpsT P1-xdh | 本研究 |
| E.coli KXW3010P3-54(P54) | E.coli W3010,pRSFdnaKJrpsT -yjhG-inv-yqhD,pEtac-kivD-inv-xdh | 本研究 |
| E.coli KXW3010P3-55(P55) | E.coli W3010,pRSFdnaKJrpsT -yjhG-inv-yqhD,pEtac-kivD-dnaKJ-xdh | 本研究 |
| E.coli KXW3010P3-56(P56) | E.coli W3010,pRSFdnaKJrpsT -yjhG-inv-yqhD,pEtac-kivD-grpE-xdh | 本研究 |
| pCas9 | P-cas9, Kanr | 实验室保存 |
| pTargetF-PMB | pTarget added sgRNA, Sper | 实验室保存 |
| pEtac-kivD-tac-xdh | Kanr, tac启动子 | 实验室保存 |
表1 本研究使用的菌株和质粒
| 菌株和质粒 | 相关特性 | 来源 |
|---|---|---|
| E.coli W3009(W3009) | E.coli W3110 ΔxylABΔyagEΔyjhHΔptsHIΔmgsA | 实验室保存 |
| E.coli W3010(W3010) | E.coli W3009 ΔyjhGΔyqhD | 本研究 |
| E.coli KXW3009(KXW3009) | E.coli W3009,pEtac-kivD-tac-xdh | 实验室保存 |
| E.coli KXW3010(KXW3010) | E.coli W3010,pEtac-kivD-tac-xdh | 本研究 |
| E.coli KXW3010P0(P0) | E.coli KXW3010,pRSFDuet-P yjhG -yjhG | 本研究 |
| E.coli KXW3010P1(P1) | E.coli KXW3010,pRSFDuet-P yjhG -yjhG-P yqhD-yqhD | 本研究 |
| E.coli KXW3010P2(P2) | E.coli KXW3010,pRSFDuet-P yjhG -yjhG-tac-yqhD | 本研究 |
| E.coli KXW3010P3(P3) | E.coli KXW3010,pRSFDuet-P yjhG -yjhG-inv-yqhD | 本研究 |
| E.coli KXW3010P4(P4) | E.coli KXW3010,pRSFDuet-P yjhG -yjhG-dnaKJ-yqhD | 本研究 |
| E.coli KXW3010P5(P5) | E.coli KXW3010,pRSFDuet-P yjhG -yjhG-grpE-yqhD | 本研究 |
| E.coli KXW3010P3-12(P12) | E.coli W3010,pRSF-P yjhG -yjhG-inv-yqhD,pEtac-kivD-rpsA P1-xdh | 本研究 |
| E.coli KXW3010P3-13(P13) | E.coli W3010,pRSF-P yjhG -yjhG-inv-yqhD,pEtac-kivD-rpsT P1-xdh | 本研究 |
| E.coli KXW3010P3-14(P14) | E.coli W3010,pRSF-P yjhG -yjhG-inv-yqhD,pEtac-kivD-inv-xdh | 本研究 |
| E.coli KXW3010P3-15(P15) | E.coli W3010,pRSF-P yjhG -yjhG-inv-yqhD,pEtac-kivD-dnaKJ-xdh | 本研究 |
| E.coli KXW3010P3-16(P16) | E.coli W3010,pRSF-P yjhG -yjhG-inv-yqhD,pEtac-kivD-grpE-xdh | 本研究 |
| E.coli KXW3010P3-21(P21) | E.coli W3010,pRSFtac-yjhG-inv-yqhD,pEtac-kivD-tac-xdh | 本研究 |
| E.coli KXW3010P3-22(P22) | E.coli W3010,pRSFtac-yjhG-inv-yqhD,pEtac-kivD-rpsA P1-xdh | 本研究 |
| E.coli KXW3010P3-23(P23) | E.coli W3010,pRSFtac-yjhG-inv-yqhD,pEtac-kivD-rpsT P1-xdh | 本研究 |
| E.coli KXW3010P3-24(P24) | E.coli W3010,pRSFtac-yjhG-inv-yqhD,pEtac-kivD-inv-xdh | 本研究 |
| E.coli KXW3010P3-25(P25) | E.coli W3010,pRSFtac-yjhG-inv-yqhD,pEtac-kivD-dnaKJ-xdh | 本研究 |
| E.coli KXW3010P3-26(P26) | E.coli W3010,pRSFtac-yjhG-inv-yqhD,pEtac-kivD-grpE-xdh | 本研究 |
| E.coli KXW3010P3-31(P31) | E.coli W3010,pRSFrrnC P1-yjhG-inv-yqhD,pEtac-kivD-tac-xdh | 本研究 |
| E.coli KXW3010P3-32(P32) | E.coli W3010,pRSFrrnC P1-yjhG-inv-yqhD,pEtac-kivD-rpsA P1-xdh | 本研究 |
| E.coli KXW3010P3-33(P33) | E.coli W3010,pRSFrrnC P1-yjhG-inv-yqhD,pEtac-kivD-rpsT P1-xdh | 本研究 |
| E.coli KXW3010P3-34(P34) | E.coli W3010,pRSFrrnC P1-yjhG-inv-yqhD,pEtac-kivD-inv-xdh | 本研究 |
| E.coli KXW3010P3-35(P35) | E.coli W3010,pRSFrrnC P1-yjhG-inv-yqhD,pEtac-kivD-dnaKJ-xdh | 本研究 |
| E.coli KXW3010P3-36(P36) | E.coli W3010,pRSFrrnC P1-yjhG-inv-yqhD,pEtac-kivD-grpE-xdh | 本研究 |
| E.coli KXW3010P3-41(P41) | E.coli W3010,pRSFrrnH P1-yjhG-inv-yqhD,pEtac-kivD-tac-xdh | 本研究 |
| E.coli KXW3010P3-42(P42) | E.coli W3010,pRSFrrnH P1-yjhG-inv-yqhD,pEtac-kivD-rpsA P1-xdh | 本研究 |
| E.coli KXW3010P3-43(P43) | E.coli W3010,pRSFrrnH P1-yjhG-inv-yqhD,pEtac-kivD-rpsT P1-xdh | 本研究 |
| E.coli KXW3010P3-44(P44) | E.coli W3010,pRSFrrnH P1-yjhG-inv-yqhD,pEtac-kivD-inv-xdh | 本研究 |
| E.coli KXW3010P3-45(P45) | E.coli W3010,pRSFrrnH P1-yjhG-inv-yqhD,pEtac-kivD-dnaKJ-xdh | 本研究 |
| E.coli KXW3010P3-46(P46) | E.coli W3010,pRSFrrnH P1-yjhG-inv-yqhD,pEtac-kivD-grpE-xdh | 本研究 |
| E.coli KXW3010P3-51(P51) | E.coli W3010,pRSFdnaKJrpsT -yjhG-inv-yqhD,pEtac-kivD-tac-xdh | 本研究 |
| E.coli KXW3010P3-52(P52) | E.coli W3010,pRSFdnaKJrpsT -yjhG-inv-yqhD,pEtac-kivD-rpsA P1-xdh | 本研究 |
| E.coli KXW3010P3-53(P53) | E.coli W3010,pRSFdnaKJrpsT -yjhG-inv-yqhD,pEtac-kivD-rpsT P1-xdh | 本研究 |
| E.coli KXW3010P3-54(P54) | E.coli W3010,pRSFdnaKJrpsT -yjhG-inv-yqhD,pEtac-kivD-inv-xdh | 本研究 |
| E.coli KXW3010P3-55(P55) | E.coli W3010,pRSFdnaKJrpsT -yjhG-inv-yqhD,pEtac-kivD-dnaKJ-xdh | 本研究 |
| E.coli KXW3010P3-56(P56) | E.coli W3010,pRSFdnaKJrpsT -yjhG-inv-yqhD,pEtac-kivD-grpE-xdh | 本研究 |
| pCas9 | P-cas9, Kanr | 实验室保存 |
| pTargetF-PMB | pTarget added sgRNA, Sper | 实验室保存 |
| pEtac-kivD-tac-xdh | Kanr, tac启动子 | 实验室保存 |
| 引物名称 | 引物序列(5′→3′) | 质粒构建或功能 |
|---|---|---|
| Sg-yjhG-F | tcaaaaaggcatcaccacccGTTTTAGAGCTAGAAATAGCAAGTTAAAATAAGGCTAGTC | pTargetF-PMB(yjhG) |
| Sg-yjhG-R | gggtggtgatgcctttttgaACTAGTATTATACCTAGGACTGAGCTAGCTGTCAAG | |
| Sg-yqhD-F | gtgatctcccgtaaaaccacGTTTTAGAGCTAGAAATAGCAAGTTAAAATAAGGCTAGTC | pTargetF-PMB(yqhD) |
| Sg-yqhD-R | gtggttttacgggagatcacACTAGTATTATACCTAGGACTGAGCTAGCTGTCAAG | |
| dornor-yjhG-F | TGGCGGGAGATCGCCAGAGCTTCCGCGCGAGCTGCGCTGAACCTATATTGCGCC | 融合片段dornor(yjhG) |
| dornor-yjhG-R | TCAATGGCGGTGGATTTGATCACCGAACCTTCTGGCGCAATATAGGTTCAGCGCAG | |
| dornor-yqhD-F | TGTGTGCTGACGCTGCCAGCAACCGGTTCAGAATCCAACGCAGGCATCACTGAAGG | 融合片段dornor(yqhD) |
| dornor-yqhD-R | GGCAATCGCGGCGTCAATACGCTCATCATCGGAACCTTCAGTGATGCCTGCGTTGG | |
| yjhG-F | aatttcacacaggaaacaGTGATCCTGTACAACTTTCCGG | pRSFDuet-P yjhG -yjhG |
| yjhG-R | cctgcaggcgcgccgGGTACCAGAAGAATTAAACCGACTTCG | |
| yqhD-F | cggcgcgcctgcaggGTCCATCAACAGTTCCTGTAACTGC | pRSFDuet-P yjhG -yjhG-P yqhD-yqhD |
| yqhD-R | tcgacttaagcattatgcAGTGGTGATGAACAGTTCTTCTCTG | |
| tac-yqhD-F | cggcgcgcctgcaggGAGGGAGCTTATCGACTGCACGG | pRSFDuet-P yjhG -yjhG-tac-yqhD |
| tac-yqhD-R | TGTTTCCTGTGTGAAATTGTTATCCGCTCAC | |
| yqhD-tac-F | aatttcacacaggaaacaGAAGTAATGAACAACTTTAATCTGCACACCCC | |
| yqhD-tac-R | tcgacttaagcattatgcGAGGCGTAAAAAGCTTAGCGGGC | |
| inv-yqhD-F | cggcgcgcctgcaggAACATTGAGGCAACGACAAGGTCGATTCTGCTATCCTTG AGGAGGCTCCTCGTAATGAACAACTTTAATCTGCACACCCC | pRSF-P yjhG -yjhG-inv-yqhD |
| inv-yqhD-R | tcgacttaagcattatgcGAGGCGTAAAAAGCTTAGCGGGC | |
| dnaKJ-yqhD-F | cggcgcgcctgcaggCAGGAAACATTTTTTGATGAATGCCTTGGC | pRSFDuet-P yjhG -yjhG-dnaKJ-yqhD |
| dnaKJ-yqhD-R | CTAAACGTCTCCACTATATATTCGGTCATCATGTGG | |
| yqhD-dnaKJ-F | cgaatatatagtggagacgtttagGTAATGAACAACTTTAATCTGCACACCCCAACC | |
| yqhD-dnaKJ-R | tcgacttaagcattatgcGAGGCGTAAAAAGCTTAGCGGGC | |
| grpE-yqhD-F | cggcgcgcctgcaggGCCTTCCAGCACATCGGCTAACTGTTGC | pRSFDuet-P yjhG -yjhG-grpE-yqhD |
| grpE-yqhD-R | GAATTTCTCCGCGTTTTTTTCGCATTCATCTC | |
| yqhD-grpE-F | gaaaaaaacgcggagaaattcGTAATGAACAACTTTAATCTGCACACCCCAACC | |
| yqhD-grpE-R | tcgacttaagcattatgcGAGGCGTAAAAAGCTTAGCGGGC | |
| rpsA P1-xdh-F | aaatcataagcggccgcaTGAAAATTTTCCTTGACGCCTCCTCGG | pEtac-kivD-rpsA P1-xdh |
| rpsA P1-xdh-R | gggatagatggctgaggacatgaaTTTGATCTCGGCCAAAAGGCGC | |
| xdh-rpsA P1-F | gccgagatcaaaTTCATGTCCTCAGCCATCTATCCCCAGCTG | |
| xdh-rpsA P1-R | tttgttagcagccggatcTCAACGCCAGCCGGCGTC | |
| rpsT P1-xdh-F | aaatcataagcggccgcaTCATTGCCATGGCGCAAATCACGG | pEtac-kivD-rpsT P1-xdh |
| rpsT P1-xdh-R | ggatagatggctgaggacatgaaACTCGTTACGTAGTGATGGCGCAG | |
| xdh-rpsT P1-F | tacgtaacgagtTTCATGTCCTCAGCCATCTATCCCCAGCTG | |
| xdh-rpsT P1-R | tttgttagcagccggatcTCAACGCCAGCCGGCGTC | |
| inv-xdh-F | taagcggccgcaAACATTGAGGCAACGACAAGGTCGATTCTGCTATCCTTG AGGAGGCTCCTCTTCATGTCCTCAGCCATCTATCCC | pEtac-kivD-inv-xdh |
| inv-xdh-R | tttgttagcagccggatcTCAACGCCAGCCGGCGTC | |
| dnaKJ-xdh-F | aaatcataagcggccgcaTTTTTTGATGAATGCCTTGGCTGCGATTCATTC | pEtac-kivD-dnaKJ-xdh |
| dnaKJ-xdh-R | CTAAACGTCTCCACTATATATTCGGTCATCATGTGGTTG | |
| xdh-dnaKJ-F | cgaatatatagtggagacgtttagGAATTCATGTCCTCAGCCATCTATCCCCAG | |
| xdh-dnaKJ-R | tttgttagcagccggatcTCAACGCCAGCCGGCGTC | |
| grpE-xdh-F | aaatcataagcggccgcaAACGTTTCTCGCTGATGTAGTGGCC | pEtac-kivD-grpE-xdh |
| grpE-xdh-R | GAATTTCTCCGCGTTTTTTTCGCATTCATCTCG | |
| xdh-grpE-F | gaaaaaaacgcggagaaattcTTCATGTCCTCAGCCATCTATCCCCAGC | |
| xdh-grpE-R | tttgttagcagccggatcTCAACGCCAGCCGGCGTC | |
| rrnC P1-yjhG-F | aatttcacacaggaaacaCTTAAAGGCATTACTTATCTTCCTTTTTCTTTTTATTCCTCC | pRSFrrnC P1-yjhG-inv-yqhD |
| rrnC P1-yjhG-R | gcaaaaatattgcgaacagacatgagTGGTGGCGCATTATAGGGAGTTATTCC | |
| yjhG-rrnC P-F | aatgcgccaccaCTCATGTCTGTTCGCAATATTTTTGCTGACG | |
| yjhG-rrnC P1-R | cctgcaggcgcgccgCTTTCAGTTTTTATTCATAAAATCGCGCAAAGCC | |
| rrnH P1-yjhG-F | aatttcacacaggaaacaAAAGTGCTGGTGCGTACGGG | pRSFrrnH P1-yjhG-inv-yqhD |
| rrnH P1-yjhG-R | caaaaatattgcgaacagacatgagTGGAGGCGCATTATAGGGAGTCGG | |
| yjhG-rrnH P-F | gcgcctccaCTCATGTCTGTTCGCAATATTTTTGCTGACG | |
| yjhG-rrnH P-R | cctgcaggcgcgccgCTTTCAGTTTTTATTCATAAAATCGCGCAAAGCC | |
| tac-yjhG-F | aatttcacacaggaaacaGGAGCTTATCGACTGCACGGTGC | pRSFtac-yjhG-inv-yqhD |
| tac-yjhG-R | TGTTTCCTGTGTGAAATTGTTATCCGCTCACAATTC | |
| yjhG-tac-F | gataacaatttcacacaggaaacaCTCATGTCTGTTCGCAATATTTTTGCTGACGAG | |
| yjhG-tac-R | cctgcaggcgcgccgCTTTCAGTTTTTATTCATAAAATCGCGCAAAGCC | |
| dnaKJ-rpsT-F | aatttcacacaggaaacaGCACAAAAAATTTTTGCATCTCCCCC | pRSFdnaKJrpsT-yjhG-inv-yqhD |
| dnaKJ-rpsT-R | ttacgtagtgatggtATTCGGTCATCATGTGGTTGTGAG | |
| rpsT-dnaKJ-F | ccacatgatgaccgaatACCATCACTACGTAACGAGTGCC | |
| rpsT-dnaKJ-R | caaaaatattgcgaacagacatgagGGTCCAACTCCCAAATGTGTTCTATATGG | |
| yjhG-KJT-F | agttggaccCTCATGTCTGTTCGCAATATTTTTGCTGACG | |
| yjhG-KJT-R | cctgcaggcgcgccgCTTTCAGTTTTTATTCATAAAATCGCGCAAAGCC |
表2 本研究使用引物
| 引物名称 | 引物序列(5′→3′) | 质粒构建或功能 |
|---|---|---|
| Sg-yjhG-F | tcaaaaaggcatcaccacccGTTTTAGAGCTAGAAATAGCAAGTTAAAATAAGGCTAGTC | pTargetF-PMB(yjhG) |
| Sg-yjhG-R | gggtggtgatgcctttttgaACTAGTATTATACCTAGGACTGAGCTAGCTGTCAAG | |
| Sg-yqhD-F | gtgatctcccgtaaaaccacGTTTTAGAGCTAGAAATAGCAAGTTAAAATAAGGCTAGTC | pTargetF-PMB(yqhD) |
| Sg-yqhD-R | gtggttttacgggagatcacACTAGTATTATACCTAGGACTGAGCTAGCTGTCAAG | |
| dornor-yjhG-F | TGGCGGGAGATCGCCAGAGCTTCCGCGCGAGCTGCGCTGAACCTATATTGCGCC | 融合片段dornor(yjhG) |
| dornor-yjhG-R | TCAATGGCGGTGGATTTGATCACCGAACCTTCTGGCGCAATATAGGTTCAGCGCAG | |
| dornor-yqhD-F | TGTGTGCTGACGCTGCCAGCAACCGGTTCAGAATCCAACGCAGGCATCACTGAAGG | 融合片段dornor(yqhD) |
| dornor-yqhD-R | GGCAATCGCGGCGTCAATACGCTCATCATCGGAACCTTCAGTGATGCCTGCGTTGG | |
| yjhG-F | aatttcacacaggaaacaGTGATCCTGTACAACTTTCCGG | pRSFDuet-P yjhG -yjhG |
| yjhG-R | cctgcaggcgcgccgGGTACCAGAAGAATTAAACCGACTTCG | |
| yqhD-F | cggcgcgcctgcaggGTCCATCAACAGTTCCTGTAACTGC | pRSFDuet-P yjhG -yjhG-P yqhD-yqhD |
| yqhD-R | tcgacttaagcattatgcAGTGGTGATGAACAGTTCTTCTCTG | |
| tac-yqhD-F | cggcgcgcctgcaggGAGGGAGCTTATCGACTGCACGG | pRSFDuet-P yjhG -yjhG-tac-yqhD |
| tac-yqhD-R | TGTTTCCTGTGTGAAATTGTTATCCGCTCAC | |
| yqhD-tac-F | aatttcacacaggaaacaGAAGTAATGAACAACTTTAATCTGCACACCCC | |
| yqhD-tac-R | tcgacttaagcattatgcGAGGCGTAAAAAGCTTAGCGGGC | |
| inv-yqhD-F | cggcgcgcctgcaggAACATTGAGGCAACGACAAGGTCGATTCTGCTATCCTTG AGGAGGCTCCTCGTAATGAACAACTTTAATCTGCACACCCC | pRSF-P yjhG -yjhG-inv-yqhD |
| inv-yqhD-R | tcgacttaagcattatgcGAGGCGTAAAAAGCTTAGCGGGC | |
| dnaKJ-yqhD-F | cggcgcgcctgcaggCAGGAAACATTTTTTGATGAATGCCTTGGC | pRSFDuet-P yjhG -yjhG-dnaKJ-yqhD |
| dnaKJ-yqhD-R | CTAAACGTCTCCACTATATATTCGGTCATCATGTGG | |
| yqhD-dnaKJ-F | cgaatatatagtggagacgtttagGTAATGAACAACTTTAATCTGCACACCCCAACC | |
| yqhD-dnaKJ-R | tcgacttaagcattatgcGAGGCGTAAAAAGCTTAGCGGGC | |
| grpE-yqhD-F | cggcgcgcctgcaggGCCTTCCAGCACATCGGCTAACTGTTGC | pRSFDuet-P yjhG -yjhG-grpE-yqhD |
| grpE-yqhD-R | GAATTTCTCCGCGTTTTTTTCGCATTCATCTC | |
| yqhD-grpE-F | gaaaaaaacgcggagaaattcGTAATGAACAACTTTAATCTGCACACCCCAACC | |
| yqhD-grpE-R | tcgacttaagcattatgcGAGGCGTAAAAAGCTTAGCGGGC | |
| rpsA P1-xdh-F | aaatcataagcggccgcaTGAAAATTTTCCTTGACGCCTCCTCGG | pEtac-kivD-rpsA P1-xdh |
| rpsA P1-xdh-R | gggatagatggctgaggacatgaaTTTGATCTCGGCCAAAAGGCGC | |
| xdh-rpsA P1-F | gccgagatcaaaTTCATGTCCTCAGCCATCTATCCCCAGCTG | |
| xdh-rpsA P1-R | tttgttagcagccggatcTCAACGCCAGCCGGCGTC | |
| rpsT P1-xdh-F | aaatcataagcggccgcaTCATTGCCATGGCGCAAATCACGG | pEtac-kivD-rpsT P1-xdh |
| rpsT P1-xdh-R | ggatagatggctgaggacatgaaACTCGTTACGTAGTGATGGCGCAG | |
| xdh-rpsT P1-F | tacgtaacgagtTTCATGTCCTCAGCCATCTATCCCCAGCTG | |
| xdh-rpsT P1-R | tttgttagcagccggatcTCAACGCCAGCCGGCGTC | |
| inv-xdh-F | taagcggccgcaAACATTGAGGCAACGACAAGGTCGATTCTGCTATCCTTG AGGAGGCTCCTCTTCATGTCCTCAGCCATCTATCCC | pEtac-kivD-inv-xdh |
| inv-xdh-R | tttgttagcagccggatcTCAACGCCAGCCGGCGTC | |
| dnaKJ-xdh-F | aaatcataagcggccgcaTTTTTTGATGAATGCCTTGGCTGCGATTCATTC | pEtac-kivD-dnaKJ-xdh |
| dnaKJ-xdh-R | CTAAACGTCTCCACTATATATTCGGTCATCATGTGGTTG | |
| xdh-dnaKJ-F | cgaatatatagtggagacgtttagGAATTCATGTCCTCAGCCATCTATCCCCAG | |
| xdh-dnaKJ-R | tttgttagcagccggatcTCAACGCCAGCCGGCGTC | |
| grpE-xdh-F | aaatcataagcggccgcaAACGTTTCTCGCTGATGTAGTGGCC | pEtac-kivD-grpE-xdh |
| grpE-xdh-R | GAATTTCTCCGCGTTTTTTTCGCATTCATCTCG | |
| xdh-grpE-F | gaaaaaaacgcggagaaattcTTCATGTCCTCAGCCATCTATCCCCAGC | |
| xdh-grpE-R | tttgttagcagccggatcTCAACGCCAGCCGGCGTC | |
| rrnC P1-yjhG-F | aatttcacacaggaaacaCTTAAAGGCATTACTTATCTTCCTTTTTCTTTTTATTCCTCC | pRSFrrnC P1-yjhG-inv-yqhD |
| rrnC P1-yjhG-R | gcaaaaatattgcgaacagacatgagTGGTGGCGCATTATAGGGAGTTATTCC | |
| yjhG-rrnC P-F | aatgcgccaccaCTCATGTCTGTTCGCAATATTTTTGCTGACG | |
| yjhG-rrnC P1-R | cctgcaggcgcgccgCTTTCAGTTTTTATTCATAAAATCGCGCAAAGCC | |
| rrnH P1-yjhG-F | aatttcacacaggaaacaAAAGTGCTGGTGCGTACGGG | pRSFrrnH P1-yjhG-inv-yqhD |
| rrnH P1-yjhG-R | caaaaatattgcgaacagacatgagTGGAGGCGCATTATAGGGAGTCGG | |
| yjhG-rrnH P-F | gcgcctccaCTCATGTCTGTTCGCAATATTTTTGCTGACG | |
| yjhG-rrnH P-R | cctgcaggcgcgccgCTTTCAGTTTTTATTCATAAAATCGCGCAAAGCC | |
| tac-yjhG-F | aatttcacacaggaaacaGGAGCTTATCGACTGCACGGTGC | pRSFtac-yjhG-inv-yqhD |
| tac-yjhG-R | TGTTTCCTGTGTGAAATTGTTATCCGCTCACAATTC | |
| yjhG-tac-F | gataacaatttcacacaggaaacaCTCATGTCTGTTCGCAATATTTTTGCTGACGAG | |
| yjhG-tac-R | cctgcaggcgcgccgCTTTCAGTTTTTATTCATAAAATCGCGCAAAGCC | |
| dnaKJ-rpsT-F | aatttcacacaggaaacaGCACAAAAAATTTTTGCATCTCCCCC | pRSFdnaKJrpsT-yjhG-inv-yqhD |
| dnaKJ-rpsT-R | ttacgtagtgatggtATTCGGTCATCATGTGGTTGTGAG | |
| rpsT-dnaKJ-F | ccacatgatgaccgaatACCATCACTACGTAACGAGTGCC | |
| rpsT-dnaKJ-R | caaaaatattgcgaacagacatgagGGTCCAACTCCCAAATGTGTTCTATATGG | |
| yjhG-KJT-F | agttggaccCTCATGTCTGTTCGCAATATTTTTGCTGACG | |
| yjhG-KJT-R | cctgcaggcgcgccgCTTTCAGTTTTTATTCATAAAATCGCGCAAAGCC |
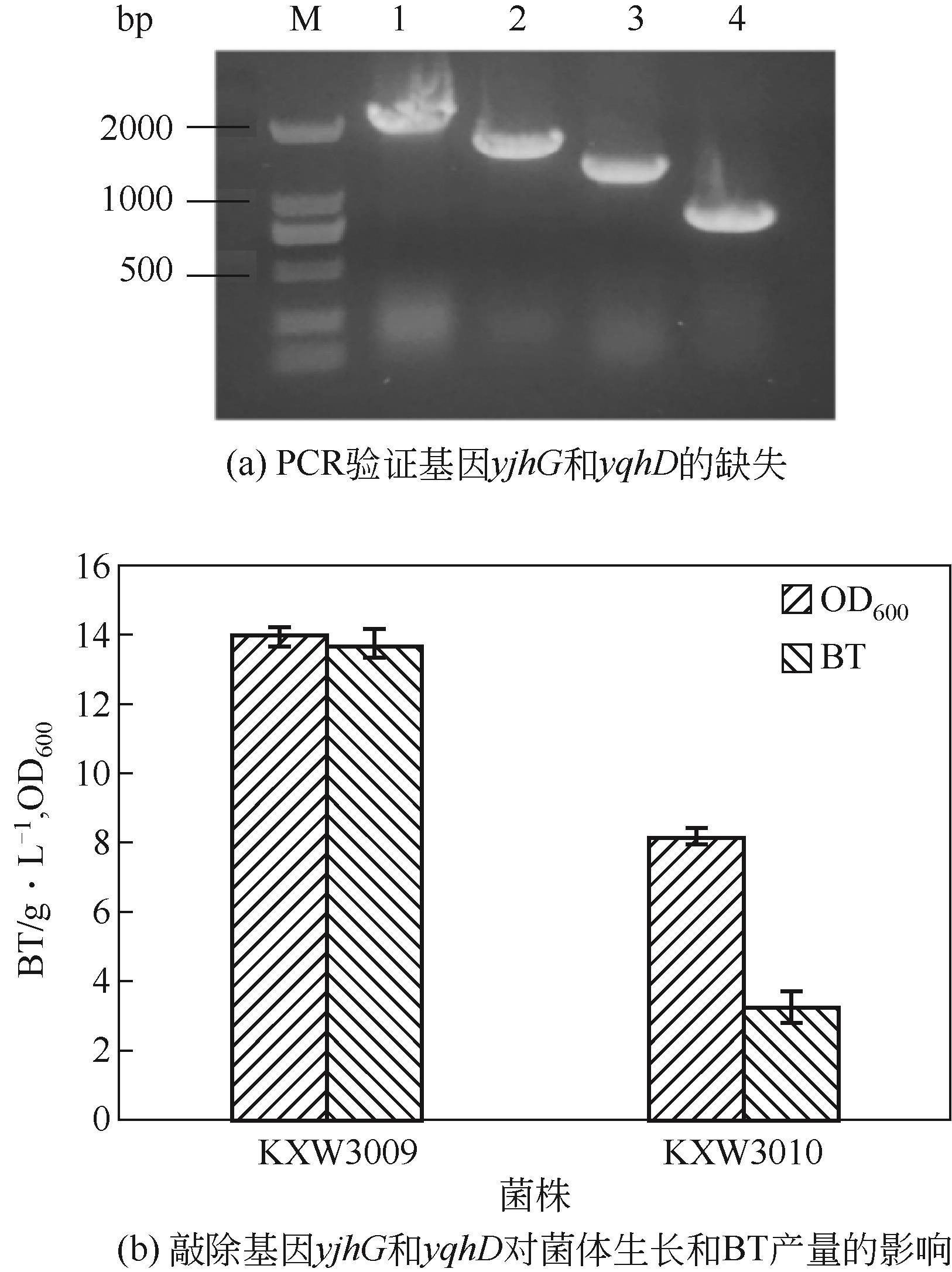
图2 敲除基因yjhG和yqhD的验证及对菌体生长和BT产量的影响M—2501 DNA marker;1—PCR验证菌株KXW3009中基因yjhG;2—PCR验证菌株KXW3010中基因yjhG;3—PCR验证菌株KXW3009中基因yqhD;4—PCR验证菌株KXW3010中基因yqhD;KXW3009—E. coli W3009, pEtac-kivD-tac-xdh;KXW3010—E. coli W3010, pEtac-kivD-tac-xdh
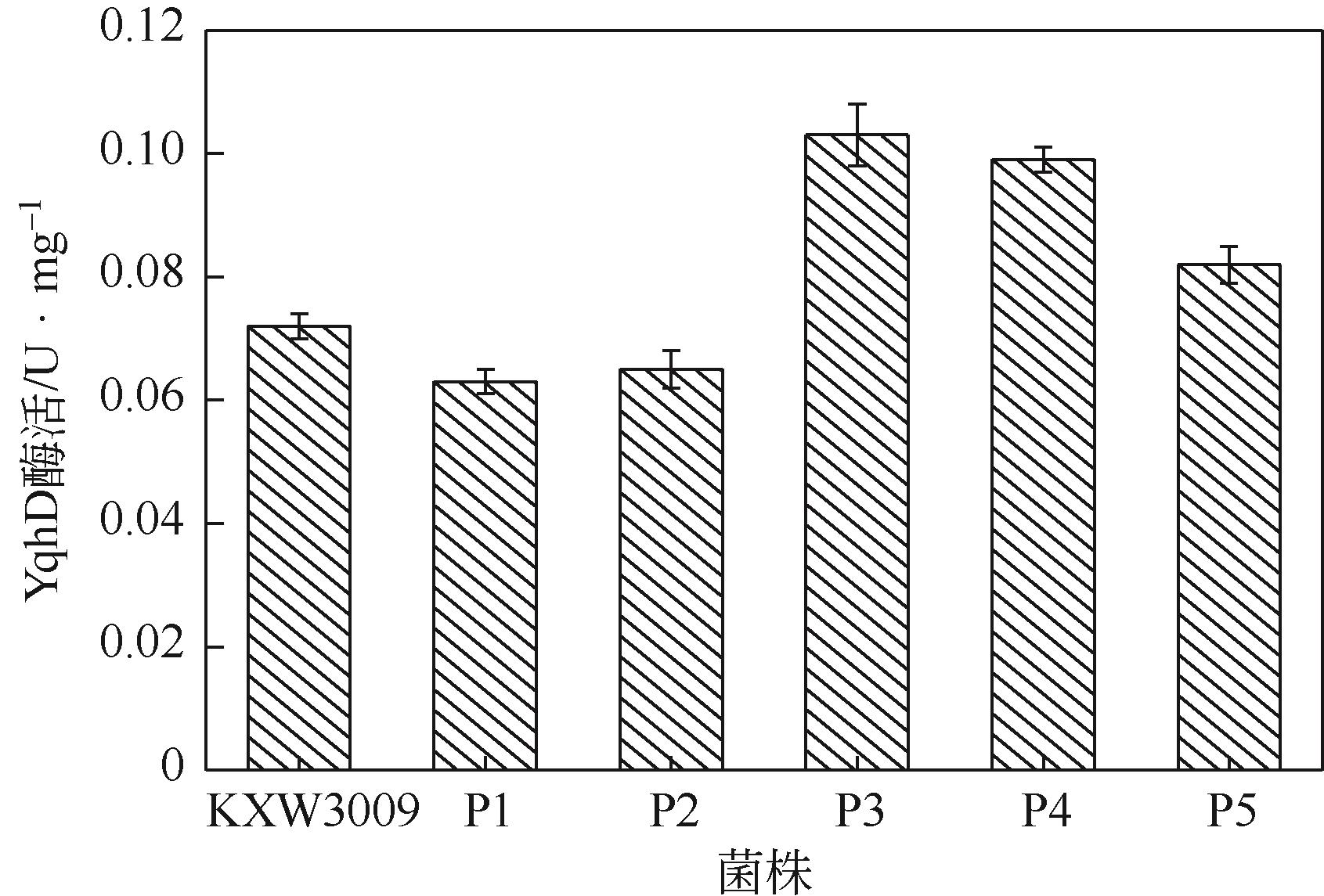
图3 重组菌中YqhD酶活菌株KXW3009—pEtac-kivD-tac-xdh;菌株P1—pRSFDuet-P yjhG -yjhG-P yqhD-yqhD;菌株P2—pRSFDuet-P yjhG -yjhG-P tac-yqhD;菌株P3—pRSFDuet-P yjhG -yjhG-P inv-yqhD;菌株P4—pRSFDuet-P yjhG -yjhG-P dnaKJ-yqhD;菌株P5—pRSFDuet-P yjhG -yjhG-P grpE-yqhD
| 菌株 | 启动子(yqhD) | OD600 | 木糖转化率/% | BT/g·L-1 |
|---|---|---|---|---|
| KXW3009 | 未改造 | 13.9 | 63.3 | 13.6 |
| P1 | P yqhD (野生型启动子) | 15.4 | 58.3 | 9.1 |
| P2 | P tac | 13.1 | 64.0 | 10.2 |
| P3 | P inv | 10.4 | 73.1 | 14.4 |
| P4 | P dnaKJ | 12.0 | 66.4 | 14.2 |
| P5 | P grpE | 11.5 | 70.0 | 14.1 |
表3 不同强度醇脱氢酶表达菌株在48 h时的发酵参数
| 菌株 | 启动子(yqhD) | OD600 | 木糖转化率/% | BT/g·L-1 |
|---|---|---|---|---|
| KXW3009 | 未改造 | 13.9 | 63.3 | 13.6 |
| P1 | P yqhD (野生型启动子) | 15.4 | 58.3 | 9.1 |
| P2 | P tac | 13.1 | 64.0 | 10.2 |
| P3 | P inv | 10.4 | 73.1 | 14.4 |
| P4 | P dnaKJ | 12.0 | 66.4 | 14.2 |
| P5 | P grpE | 11.5 | 70.0 | 14.1 |
| 菌株 | 启动子(xdh) | 启动子(yjhG) | BT/g·L-1 | OD600 | 木糖消耗率/% | 菌株 | 启动子(xdh) | 启动子(yjhG) | BT/g·L-1 | OD600 | 木糖消耗率/% |
|---|---|---|---|---|---|---|---|---|---|---|---|
| P3 | P tac | P yjhG | 14.1 | 12.2 | 63.8 | P34 | P inv | P rrnC P1 | 3.2 | 8.0 | 22.4 |
| P12 | P rpsA P1 | P yjhG | 9.2 | 9.9 | 44.5 | P35 | P dnaKJ | P rrnC P1 | 6.7 | 14.3 | 36.7 |
| P13 | P rpsT P1 | P yjhG | 9.6 | 11.1 | 40.8 | P36 | P grpE | P rrnC P1 | 5.1 | 11.6 | 40.9 |
| P14 | P inv | P yjhG | 11.4 | 9.2 | 47.9 | P41 | P tac | P rrnH P1 | 15.6 | 12.3 | 65.7 |
| P15 | P dnaKJ | P yjhG | 10.1 | 11.4 | 40.4 | P42 | P rpsA P1 | P rrnH P1 | 7.7 | 10.6 | 24.4 |
| P16 | P grpE | P yjhG | 5.8 | 11.8 | 35.9 | P43 | P rpsT P1 | P rrnH P1 | 8.1 | 10.8 | 25.0 |
| P21 | P tac | P tac | 7.6 | 12.0 | 47.3 | P44 | P inv | P rrnH P1 | 10.2 | 9.6 | 48.1 |
| P22 | P rpsA P1 | P tac | 8.2 | 9.5 | 24.6 | P45 | P dnaKJ | P rrnH P1 | 8.0 | 13.4 | 28.8 |
| P23 | P rpsT P1 | P tac | 4.4 | 11.2 | 19.6 | P46 | P grpE | P rrnH P1 | 5.23 | 10.5 | 34.2 |
| P24 | P inv | P tac | 11.1 | 9.2 | 47.7 | P51 | P tac | P dnaKJrpsT | 9.6 | 12.7 | 43.0 |
| P25 | P dnaKJ | P tac | 7.4 | 12.1 | 27.2 | P52 | P rpsA P1 | P dnaKJrpsT | 6.2 | 9.4 | 24.0 |
| P26 | P grpE | P tac | 5.0 | 11.1 | 33.5 | P53 | P rpsT P1 | P dnaKJrpsT | 5.7 | 11.7 | 21.9 |
| P31 | P tac | P rrnC P1 | 6.4 | 12.9 | 29.3 | P54 | P inv | P dnaKJrpsT | 2.9 | 13.0 | 17.0 |
| P32 | P rpsA P1 | P rrnC P1 | 6.7 | 9.6 | 30.4 | P55 | P dnaKJ | P dnaKJrpsT | 7.8 | 12.2 | 25.2 |
| P33 | P rpsT P1 | P rrnC P1 | 5.2 | 12.4 | 35.9 | P56 | P grpE | P dnaKJrpsT | 5.7 | 10.8 | 35.9 |
表4 不同菌株在48h时的发酵参数
| 菌株 | 启动子(xdh) | 启动子(yjhG) | BT/g·L-1 | OD600 | 木糖消耗率/% | 菌株 | 启动子(xdh) | 启动子(yjhG) | BT/g·L-1 | OD600 | 木糖消耗率/% |
|---|---|---|---|---|---|---|---|---|---|---|---|
| P3 | P tac | P yjhG | 14.1 | 12.2 | 63.8 | P34 | P inv | P rrnC P1 | 3.2 | 8.0 | 22.4 |
| P12 | P rpsA P1 | P yjhG | 9.2 | 9.9 | 44.5 | P35 | P dnaKJ | P rrnC P1 | 6.7 | 14.3 | 36.7 |
| P13 | P rpsT P1 | P yjhG | 9.6 | 11.1 | 40.8 | P36 | P grpE | P rrnC P1 | 5.1 | 11.6 | 40.9 |
| P14 | P inv | P yjhG | 11.4 | 9.2 | 47.9 | P41 | P tac | P rrnH P1 | 15.6 | 12.3 | 65.7 |
| P15 | P dnaKJ | P yjhG | 10.1 | 11.4 | 40.4 | P42 | P rpsA P1 | P rrnH P1 | 7.7 | 10.6 | 24.4 |
| P16 | P grpE | P yjhG | 5.8 | 11.8 | 35.9 | P43 | P rpsT P1 | P rrnH P1 | 8.1 | 10.8 | 25.0 |
| P21 | P tac | P tac | 7.6 | 12.0 | 47.3 | P44 | P inv | P rrnH P1 | 10.2 | 9.6 | 48.1 |
| P22 | P rpsA P1 | P tac | 8.2 | 9.5 | 24.6 | P45 | P dnaKJ | P rrnH P1 | 8.0 | 13.4 | 28.8 |
| P23 | P rpsT P1 | P tac | 4.4 | 11.2 | 19.6 | P46 | P grpE | P rrnH P1 | 5.23 | 10.5 | 34.2 |
| P24 | P inv | P tac | 11.1 | 9.2 | 47.7 | P51 | P tac | P dnaKJrpsT | 9.6 | 12.7 | 43.0 |
| P25 | P dnaKJ | P tac | 7.4 | 12.1 | 27.2 | P52 | P rpsA P1 | P dnaKJrpsT | 6.2 | 9.4 | 24.0 |
| P26 | P grpE | P tac | 5.0 | 11.1 | 33.5 | P53 | P rpsT P1 | P dnaKJrpsT | 5.7 | 11.7 | 21.9 |
| P31 | P tac | P rrnC P1 | 6.4 | 12.9 | 29.3 | P54 | P inv | P dnaKJrpsT | 2.9 | 13.0 | 17.0 |
| P32 | P rpsA P1 | P rrnC P1 | 6.7 | 9.6 | 30.4 | P55 | P dnaKJ | P dnaKJrpsT | 7.8 | 12.2 | 25.2 |
| P33 | P rpsT P1 | P rrnC P1 | 5.2 | 12.4 | 35.9 | P56 | P grpE | P dnaKJrpsT | 5.7 | 10.8 | 35.9 |
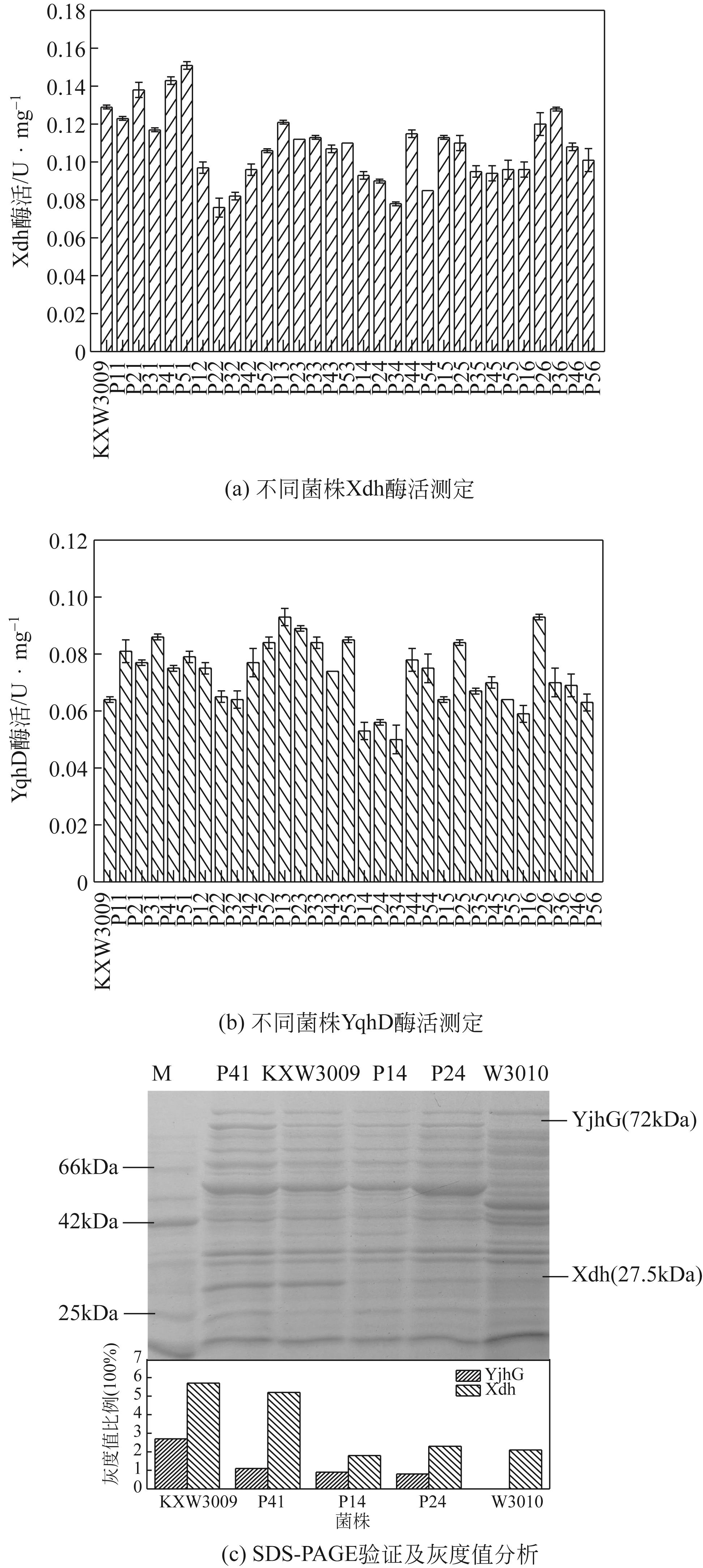
图5 重组菌中Xdh和YqhD的酶活及SDS-PAGE电泳分析M—蛋白marker;菌株KXW3009—pEtac-kivD-tac-xdh;菌株P41—pRSFrrnH P1-yjhG-inv-yqhD, pEtac-kivD-tac-xdh;菌株P24—pRSFtac-yjhG-inv-yqhD, pEtac-kivD-inv-xdh;菌株P14—pRSF-P yjhG -yjhG-inv-yqhD, pEtac-kivD-inv-xdh;菌株W3010—菌株KXW3009 ΔyjhGΔyqhD
| 1 | NIU W, MOLEFE M N, FROST J W. Microbial synthesis of the energetic material precursor 1,2,4-butanetriol[J]. Journal of the American Chemical Society, 2003, 125(43): 12998-12999. |
| 2 | REN T, LIU D X. Synthesis of cationic lipids from 1, 2, 4-butanetriol[J]. Tetrahedron Letters, 1999, 40(2): 209-212. |
| 3 | 景培源. 多策略强化1,2,4-丁三醇的生物合成[D]. 无锡: 江南大学, 2018. |
| JING Peiyuan. Enhanced 1,2,4-butanetriol bioproduction by muti-strategy [D]. Wuxi: Jiangnan University, 2018. | |
| 4 | 何姝颖, 诸葛斌, 陆信曜, 等. 副产物途径的缺失对大肠杆菌合成D-1, 2, 4-丁三醇的影响[J]. 微生物学通报, 2017, 44(1): 30-37. |
| HE S Y, ZHUGE B, LU X Y, et al. Influence of the deficiency of by-product pathways on biosynthesis of D-1,2,4-butanetriol in Escherichia coli [J]. Microbiology China, 2017, 44(1): 30-37. | |
| 5 | 祖金珊, 徐世文, 陆信曜, 等. 整合型1, 2, 4-丁三醇重组菌的构建及共底物发酵[J]. 应用与环境生物学报, 2019, 25(4): 966-971. |
| ZU J S, XU S W, LU X Y, et al. Construction of a 1, 2, 4-butanetriol-integrated strain and fermentation of its co-substrate[J]. Chinese Journal of Applied and Environmental Biology, 2019, 25(4): 966-971. | |
| 6 | VALDEHUESA K N G, LIU H W, RAMOS K R M, et al. Direct bioconversion of d-xylose to 1, 2, 4-butanetriol in an engineered Escherichia coli [J]. Process Biochemistry, 2014, 49(1): 25-32. |
| 7 | 孙雷, 杨帆, 朱泰承, 等. 大肠杆菌合成1,2,4-丁三醇的途径优化[J]. 生物工程学报, 2016, 32(1): 51-63. |
| SUN L, YANG F, ZHU T C, et al. Optimization of 1,2,4-butanetriol synthetic pathway in Escherichia coli [J]. Chinese Journal of Biotechnology, 2016, 32(1): 51-63. | |
| 8 | ZHOU S H, YUAN S F, NAIR P H, et al. Development of a growth coupled and multi-layered dynamic regulation network balancing malonyl-CoA node to enhance (2S)-naringenin biosynthesis in Escherichia coli [J]. Metabolic Engineering, 2021, 67: 41-52. |
| 9 | CABULONG R B, VALDEHUESA K N G, RAMOS K R M, et al. Enhanced yield of ethylene glycol production from D-xylose by pathway optimization in Escherichia coli [J]. Enzyme and Microbial Technology, 2017, 97: 11-20. |
| 10 | MA Q, XIA L, WU H Y, et al. Metabolic engineering of Escherichia coli for efficient osmotic stress-free production of compatible solute hydroxyectoine[J]. Biotechnology and Bioengineering, 2022, 119(1): 89-101. |
| 11 | HOU J S, GAO C, GUO L, et al. Rewiring carbon flux in Escherichia coli using a bifunctional molecular switch[J]. Metabolic Engineering, 2020, 61: 47-57. |
| 12 | GAO C, HOU J S, XU P, et al. Programmable biomolecular switches for rewiring flux in Escherichia coli [J]. Nature Communications, 2019, 10(1): 3751. |
| 13 | 韦宇拓, 黄日波, 杜丽琴.一个在大肠杆菌中组成型表达的启动子及其在大肠杆菌中的应用: CN101560516B[P]. 1970-01-16. |
| WEI Y T, HUANG R B, DU L Q. Constitutive expression promoter in Escherichia coli and applications in Escherichia coli : CN101560516B[P]. 1970-01-16. | |
| 14 | 周景文, 陈坚, 周胜虎, 等. 一种大肠杆菌高强度组合启动子及其应用: CN106591310B[P]. 2019-10-25. |
| ZHOU J W, CHEN J, ZHOU S H, et al. Escherichia coli high-strength combined promoters and application thereof: CN106591310B[P]. 2019-10-25. | |
| 15 | MAEDA M, SHIMADA T, ISHIHAMA A. Strength and regulation of seven rRNA promoters in Escherichia coli [J]. PLoS One, 2015, 10(12): e0144697. |
| 16 | 狄莹莹, 王昕钰, 冯奥, 等. 大肠杆菌乙酸弱化对1,2,4-丁三醇合成的影响[J].食品与发酵工业, 2021, 47(19): 29-34. |
| DI Y Y, WANG X Y, FENG A, et al. Improved 1,2,4-butanetriol production by weaken acetate pathway in a recombinant Escherichia coli [J]. Food and Fermentation Industries, 2021, 47(19): 29-34. | |
| 17 | ATSUMI S, WU T Y, ECKL E M, et al. Engineering the isobutanol biosynthetic pathway in Escherichia coli by comparison of three aldehyde reductase/alcohol dehydrogenase genes[J]. Applied Microbiology and Biotechnology, 2010, 85(3): 651-657. |
| 18 | 朱晓, 邹琪琪, 张锋. 拜氏梭菌(Clostridium beijerinckii)NRRL-B593在三种不同培养基中乙醇脱氢酶活力的测定[J]. 科技风, 2018(15): 130-131. |
| ZHU X, ZOU Q Q, ZHANG F. Determination of alcohol dehydrogenase activities in Clostridium beijerinckii grown on various substrates[J]. Technology Wind, 2018(15): 130-131. | |
| 19 | ZHANG Y P, GUO S T, WANG Y X, et al. Production of D-xylonate from corn cob hydrolysate by a metabolically engineered Escherichia coli strain[J]. ACS Sustainable Chemistry & Engineering, 2019, 7(2): 2160-2168. |
| 20 | VALDEHUESA K N G, LEE W K, RAMOS K R M, et al. Identification of aldehyde reductase catalyzing the terminal step for conversion of xylose to butanetriol in engineered Escherichia coli [J]. Bioprocess and Biosystems Engineering, 2015, 38(9): 1761-1772. |
| 21 | SUN L, YANG F, SUN H B, et al. Synthetic pathway optimization for improved 1,2,4-butanetriol production[J]. Journal of Industrial Microbiology and Biotechnology, 2016, 43(1): 67-78. |
| 22 | ZHANG N N, WANG J B, ZHANG Y, et al. Metabolic pathway optimization for biosynthesis of 1,2,4-butanetriol from xylose by engineered Escherichia coli [J]. Enzyme and Microbial Technology, 2016, 93/94: 51-58. |
| 23 | 祖金珊. 整合型1,2,4-丁三醇合成菌株的构建及发酵条件优化[D]. 无锡: 江南大学, 2019. |
| ZU J S. Construction of integrated 1,2,4-butanetriol synthesis strain and optimization of fermentation conditions[D]. Wuxi: Jiangnan University, 2019. | |
| 24 | TOIVARI M, VEHKOMÄKI M L, NYGÅRD Y, et al. Low pH D-xylonate production with Pichia kudriavzevii [J]. Bioresource Technology, 2013, 133: 555-562. |
| 25 | ANGLÈS F, CASTANIÉ-CORNET M P, SLAMA N, et al. Multilevel interaction of the DnaK/DnaJ(HSP70/HSP40) stress-responsive chaperone machine with the central metabolism[J]. Scientific Reports, 2017, 7: 41341. |
| 26 | DARIUSHNEJAD H, FARAJNIA S, ZARGHAMI N, et al. Effect of DnaK/DnaJ/GrpE and DsbC chaperons on periplasmic expression of Fab antibody by E. coli SEC pathway[J]. International Journal of Peptide Research and Therapeutics, 2019, 25(1): 67-74. |
| [1] | 唐文秀, 王学明, 郭亮, 季立豪, 高聪, 陈修来, 刘立明. 代谢工程改造大肠杆菌生产琥珀酸[J]. 化工进展, 2022, 41(2): 938-950. |
| [2] | 陶雨萱, 张尚杰, 景艺文, 信丰学, 董维亮, 周杰, 蒋羽佳, 章文明, 姜岷. 甲基营养型大肠杆菌构建策略的研究进展[J]. 化工进展, 2021, 40(7): 3932-3941. |
| [3] | 李阳, 朱晨辉, 范代娣. 重组胶原蛋白的绿色生物制造及其应用[J]. 化工进展, 2021, 40(3): 1262-1275. |
| [4] | 陈露,刘丁玉,汪保卫,赵玉姣,贾广韬,陈涛,王智文. 大肠杆菌乙酰辅酶A代谢调控及其应用研究进展[J]. 化工进展, 2019, 38(9): 4218-4226. |
| [5] | 宋鑫, 张双虹, 陈涛, 刘训理, 王智文. 大肠杆菌调控因子工程及其菌株耐受性研究进展[J]. 化工进展, 2018, 37(07): 2780-2789. |
| [6] | 吕蒙蒙, 刘蛟, 刘欢欢, 陈红, 王成, 闻建平. 平衡磷酸烯醇式丙酮酸节点通量强化筑波链霉菌合成他克莫司[J]. 化工进展, 2018, 37(06): 2347-2353. |
| [7] | 于化江1,熊 亮2,熊中琼2,张国庆2. 复合电沉积制备TiO2/泡沫镍光催化材料及其催化活性 [J]. 化工进展, 2011, 30(9): 1972-. |
| [8] | 陈文谋,卢英华,李清彪,程志敬,何 宁 . 固定床反应器培养固定化大肠杆菌生产β-葡聚糖酶 [J]. 化工进展, 2006, 25(12): 1423-. |
| [9] | 聂 尧,徐 岩,王海燕,许 娜,肖 荣. 重组大肠杆菌不对称还原2-羟基苯乙酮合成(R)-苯基乙二醇 [J]. 化工进展, 2006, 25(10): 1231-. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||

