Chemical Industry and Engineering Progress ›› 2021, Vol. 40 ›› Issue (7): 3932-3941.DOI: 10.16085/j.issn.1000-6613.2020-1607
• Biochemical and pharmaceutical engineering • Previous Articles Next Articles
Recent advances in the construction strategy of methylotrophic Escherichia coli
TAO Yuxuan1( ), ZHANG Shangjie1, JING Yiwen1, XIN Fengxue1,2, DONG Weiliang1,2, ZHOU Jie1,2, JIANG Yujia1, ZHANG Wenming1,2(
), ZHANG Shangjie1, JING Yiwen1, XIN Fengxue1,2, DONG Weiliang1,2, ZHOU Jie1,2, JIANG Yujia1, ZHANG Wenming1,2( ), JIANG Min1,2(
), JIANG Min1,2( )
)
- 1.State Key Laboratory of Materials-Oriented Chemical Engineering, College of Biotechnology and Pharmaceutical Engineering, Nanjing Tech University, Nanjing 211816, Jiangsu, China
2.Jiangsu National Synergetic Innovation Center for Advanced Materials (SICAM), Nanjing Tech University, Nanjing 211816, Jiangsu, China
-
Received:2020-08-12Revised:2020-10-26Online:2021-07-19Published:2021-07-06 -
Contact:ZHANG Wenming,JIANG Min
甲基营养型大肠杆菌构建策略的研究进展
陶雨萱1( ), 张尚杰1, 景艺文1, 信丰学1,2, 董维亮1,2, 周杰1,2, 蒋羽佳1, 章文明1,2(
), 张尚杰1, 景艺文1, 信丰学1,2, 董维亮1,2, 周杰1,2, 蒋羽佳1, 章文明1,2( ), 姜岷1,2(
), 姜岷1,2( )
)
- 1.南京工业大学生物与制药工程学院, 材料化学工程国家重点实验室,江苏 南京 211816
2.南京工业大学, 江苏先进生物与化学制造协同创新中心(SICAM),江苏 南京 211816
-
通讯作者:章文明,姜岷 -
作者简介:陶雨萱(1997—),女,硕士研究生,主要研究方向为抗逆元件的挖掘与性能表征。E-mail:15106192105@163.com 。 -
基金资助:国家重点研发计划(2018YFA0901500);国家自然科学基金(22078151)
CLC Number:
Cite this article
TAO Yuxuan, ZHANG Shangjie, JING Yiwen, XIN Fengxue, DONG Weiliang, ZHOU Jie, JIANG Yujia, ZHANG Wenming, JIANG Min. Recent advances in the construction strategy of methylotrophic Escherichia coli[J]. Chemical Industry and Engineering Progress, 2021, 40(7): 3932-3941.
陶雨萱, 张尚杰, 景艺文, 信丰学, 董维亮, 周杰, 蒋羽佳, 章文明, 姜岷. 甲基营养型大肠杆菌构建策略的研究进展[J]. 化工进展, 2021, 40(7): 3932-3941.
share this article
Add to citation manager EndNote|Ris|BibTeX
URL: https://hgjz.cip.com.cn/EN/10.16085/j.issn.1000-6613.2020-1607
| 底物 | 宿主 | 改造方法 | 改造结果 | 文献 |
|---|---|---|---|---|
| 酵母提取物+甲醇 | E. coli ΔfrmA | 引入甲醇芽孢杆菌RuMP途径酶 表达了密码子优化的甲醇芽孢杆菌HPS和PHI基因 | 甲醛消耗量从(0.058±0.013)mm增加到(0.074±0.001)mm | [ |
| 甲醇 | E. coliΔfrmA | 利用SH3-配体组装工程超分子酶复合物 | 甲醇转化为F6P的能力提高了50倍 | [ |
| 甲醇+氨基酸 | E. coliΔlrp | 苏氨酸共利用 敲除亮氨酸反馈调节蛋白(LRP) | 利用酵母提取物和甲醇使生物量最终浓度增加33% Δlrp菌株的生物量在有甲醇的情况下比不加甲醇的高34% | [ |
| 甲醇+葡萄糖酸盐 | MeSV1 | 敲除磷酸葡萄糖酸脱水酶(EDD)和5-磷酸核糖异构酶(RPIAB) | 高达24%的甲醇进入中心代谢 不加酵母粉培养的MeSV2.1 第九代最大生长量达到OD600=1.34±0.03 | [ |
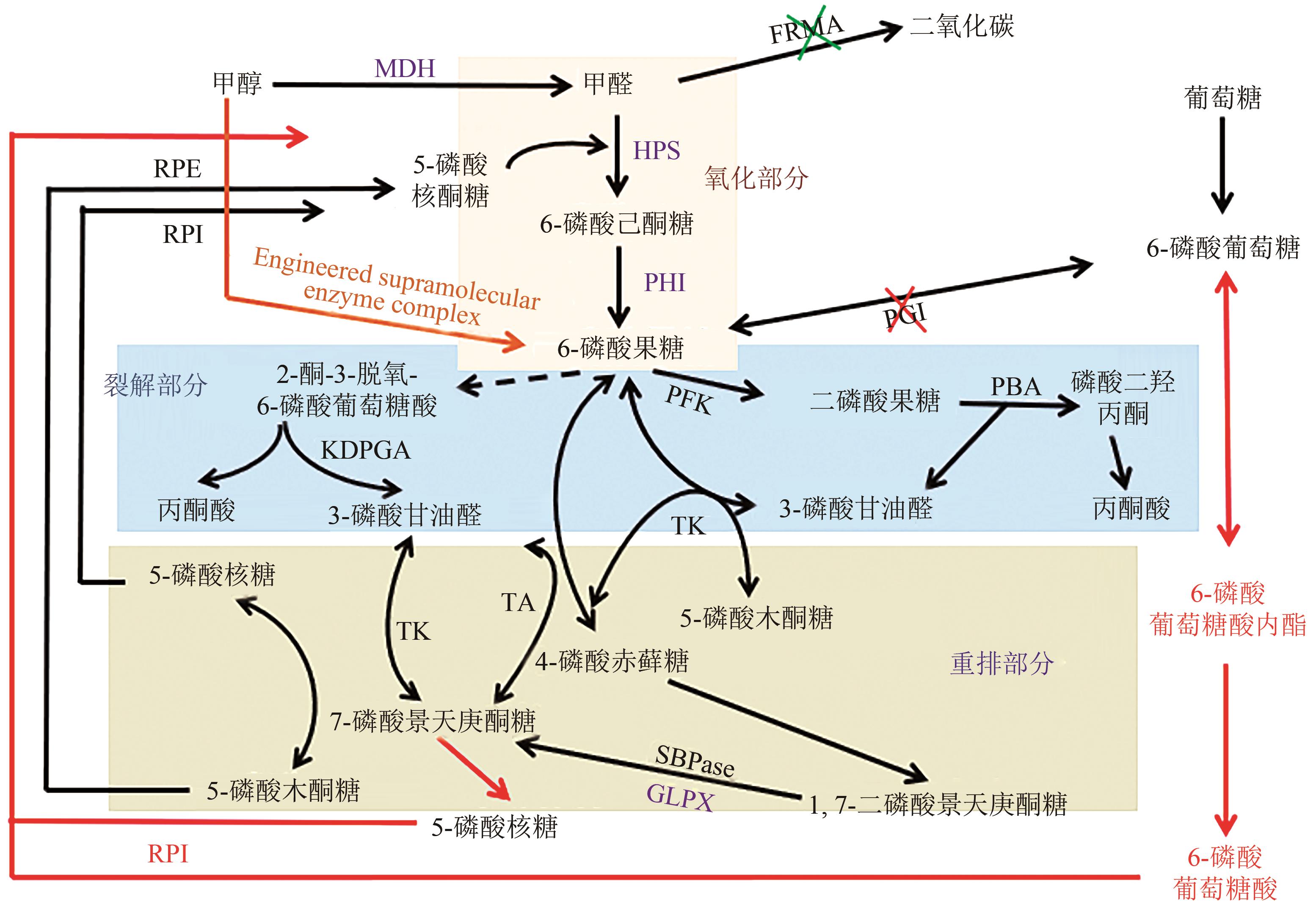
| 甲醇+葡萄糖 | E. coliΔfrmA | 表达甲醇芽孢杆菌的PPP途径 敲除磷酸葡萄糖异构酶基因(PGI)和甲醛脱氢酶基因(FRMA) | 甲基营养大肠杆菌ΔfrmAΔpgi的甲醇利用率比甲基营养大肠杆菌ΔfrmA提高了219% | [ |
| 甲醇+木糖 | E. coli | 碘乙酸酯抑制糖酵解通量 过量表达景天庚酮糖二磷酸酶(GLPX) | Ru5P相应增加(4.0±1.2)倍,甲醛降幅为2.50%±0.7% | [ |
| 甲醇 | E. coli | 拷贝数变异(CNVs) 适应性进化 | 构建菌株能够以甲醇为唯一碳源生长 在8h的倍增时间内高效生长 | [ |
| 甲醇+葡萄糖 | E. coli | 敲除葡萄糖6-磷酸异构酶(PGI)、磷酸葡萄糖酸脱水酶(EDD)和核糖5-磷酸异构酶(RPIAB) | 进化菌株在含甲醇的葡萄糖微量培养基中的最大生长速率为0.15/h | [ |
| 甲醇+木糖 | E. coli | 将菌株对甲醇的利用与在五碳(C5)糖上的生长相结合 敲除用于戊糖利用的戊糖磷酸途径中的必要基因并表达来自RuMP途径的异源酶 | 菌株能够以(0.17±0.006)h-1的速率利用甲醇,甲醇和木糖以大约1∶1的摩尔比共同化 | [ |
| 底物 | 宿主 | 改造方法 | 改造结果 | 文献 |
|---|---|---|---|---|
| 酵母提取物+甲醇 | E. coli ΔfrmA | 引入甲醇芽孢杆菌RuMP途径酶 表达了密码子优化的甲醇芽孢杆菌HPS和PHI基因 | 甲醛消耗量从(0.058±0.013)mm增加到(0.074±0.001)mm | [ |
| 甲醇 | E. coliΔfrmA | 利用SH3-配体组装工程超分子酶复合物 | 甲醇转化为F6P的能力提高了50倍 | [ |
| 甲醇+氨基酸 | E. coliΔlrp | 苏氨酸共利用 敲除亮氨酸反馈调节蛋白(LRP) | 利用酵母提取物和甲醇使生物量最终浓度增加33% Δlrp菌株的生物量在有甲醇的情况下比不加甲醇的高34% | [ |
| 甲醇+葡萄糖酸盐 | MeSV1 | 敲除磷酸葡萄糖酸脱水酶(EDD)和5-磷酸核糖异构酶(RPIAB) | 高达24%的甲醇进入中心代谢 不加酵母粉培养的MeSV2.1 第九代最大生长量达到OD600=1.34±0.03 | [ |
| 甲醇+葡萄糖 | E. coliΔfrmA | 表达甲醇芽孢杆菌的PPP途径 敲除磷酸葡萄糖异构酶基因(PGI)和甲醛脱氢酶基因(FRMA) | 甲基营养大肠杆菌ΔfrmAΔpgi的甲醇利用率比甲基营养大肠杆菌ΔfrmA提高了219% | [ |
| 甲醇+木糖 | E. coli | 碘乙酸酯抑制糖酵解通量 过量表达景天庚酮糖二磷酸酶(GLPX) | Ru5P相应增加(4.0±1.2)倍,甲醛降幅为2.50%±0.7% | [ |
| 甲醇 | E. coli | 拷贝数变异(CNVs) 适应性进化 | 构建菌株能够以甲醇为唯一碳源生长 在8h的倍增时间内高效生长 | [ |
| 甲醇+葡萄糖 | E. coli | 敲除葡萄糖6-磷酸异构酶(PGI)、磷酸葡萄糖酸脱水酶(EDD)和核糖5-磷酸异构酶(RPIAB) | 进化菌株在含甲醇的葡萄糖微量培养基中的最大生长速率为0.15/h | [ |
| 甲醇+木糖 | E. coli | 将菌株对甲醇的利用与在五碳(C5)糖上的生长相结合 敲除用于戊糖利用的戊糖磷酸途径中的必要基因并表达来自RuMP途径的异源酶 | 菌株能够以(0.17±0.006)h-1的速率利用甲醇,甲醇和木糖以大约1∶1的摩尔比共同化 | [ |
| 底物 | 宿主 | 改造方法 | 改造结果 | 文献 |
|---|---|---|---|---|
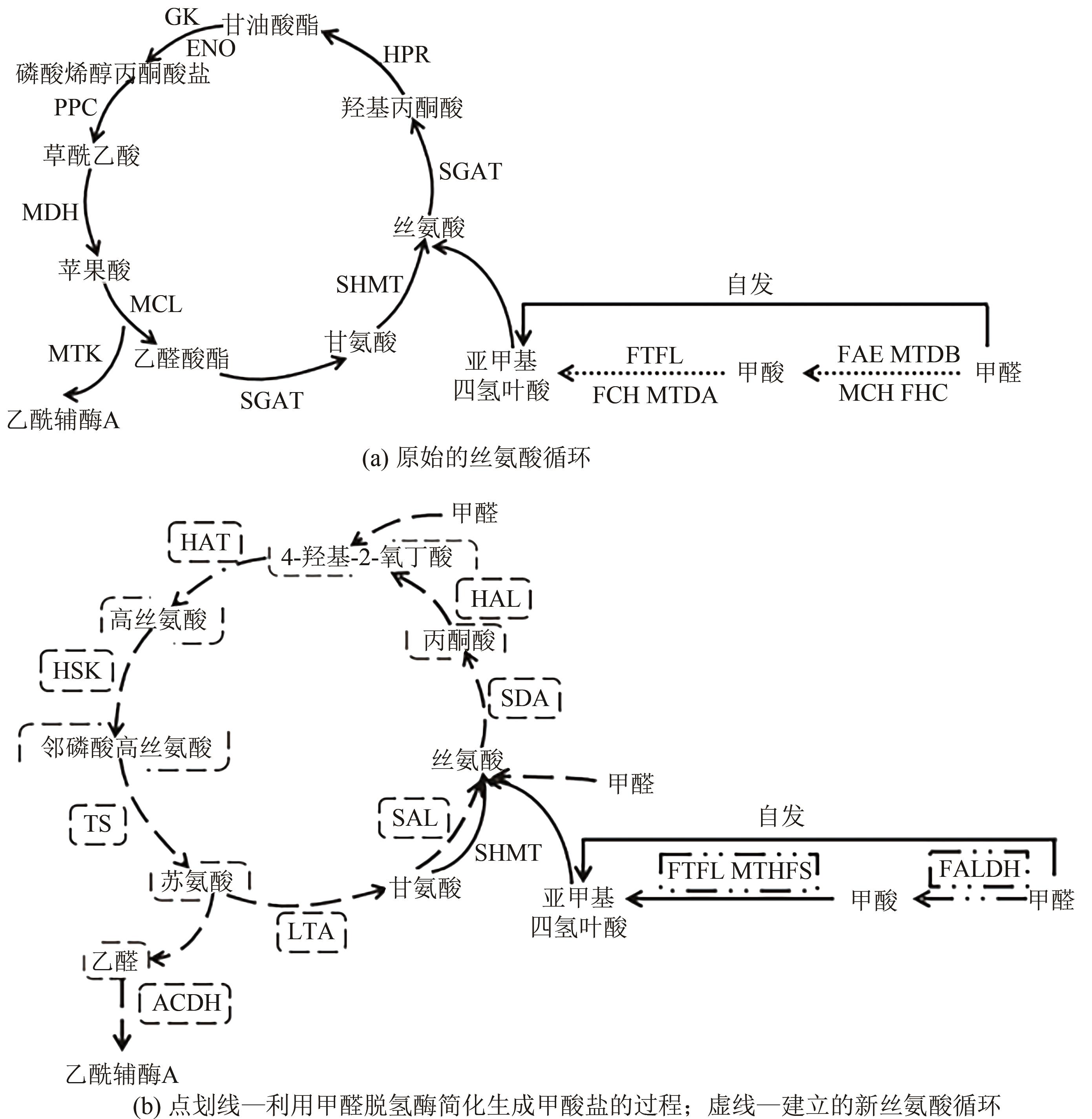
| 木糖+甲醇+碳酸氢盐 | E. coliΔserA ΔgcvP | 改进丝氨酸循环 使用甲醛脱氢酶(FALDH)来简化甲醛氧化成甲酸盐的过程 | 最快的甲醇同化速率为0.7mm·h-1(OD600),产生乙醇约6.7mmol·L-1 | [ |
| 甲酸+葡萄糖 | E. coli strain DH5α | 合成丝氨酸-苏氨酸循环 | 证明甲酸盐可以作为细胞碳一的唯一来源(倍增时间为2.3h) | [ |
| 甲醇+葡萄糖 | E. coliMG1655 | 构建高丝氨酸循环 | 生物量产量比丝氨酸循环高13% | [ |
| 底物 | 宿主 | 改造方法 | 改造结果 | 文献 |
|---|---|---|---|---|
| 木糖+甲醇+碳酸氢盐 | E. coliΔserA ΔgcvP | 改进丝氨酸循环 使用甲醛脱氢酶(FALDH)来简化甲醛氧化成甲酸盐的过程 | 最快的甲醇同化速率为0.7mm·h-1(OD600),产生乙醇约6.7mmol·L-1 | [ |
| 甲酸+葡萄糖 | E. coli strain DH5α | 合成丝氨酸-苏氨酸循环 | 证明甲酸盐可以作为细胞碳一的唯一来源(倍增时间为2.3h) | [ |
| 甲醇+葡萄糖 | E. coliMG1655 | 构建高丝氨酸循环 | 生物量产量比丝氨酸循环高13% | [ |
| 底物 | 宿主 | 改造方法 | 改造结果 | 文献 |
|---|---|---|---|---|
| 甲酸盐 | fdoG MG1655-derived E. coli strain | 计算机设计甲醛酶(FLS)实现碳一到碳三化合物的合成 | 很高的化学推动力(>3kcal·mol-1,1cal=4.1840J) FLS活性增加了100倍 | [ |
| 甲醇+酵母膏 | BW25113frmA(EC-ΔfrmA) | 高效表达MDH和人工FLS 组装人工线性甲醇同化途径 | 甲醇消耗量从原始1g·L-1增加到1.45g·L-1 | [ |
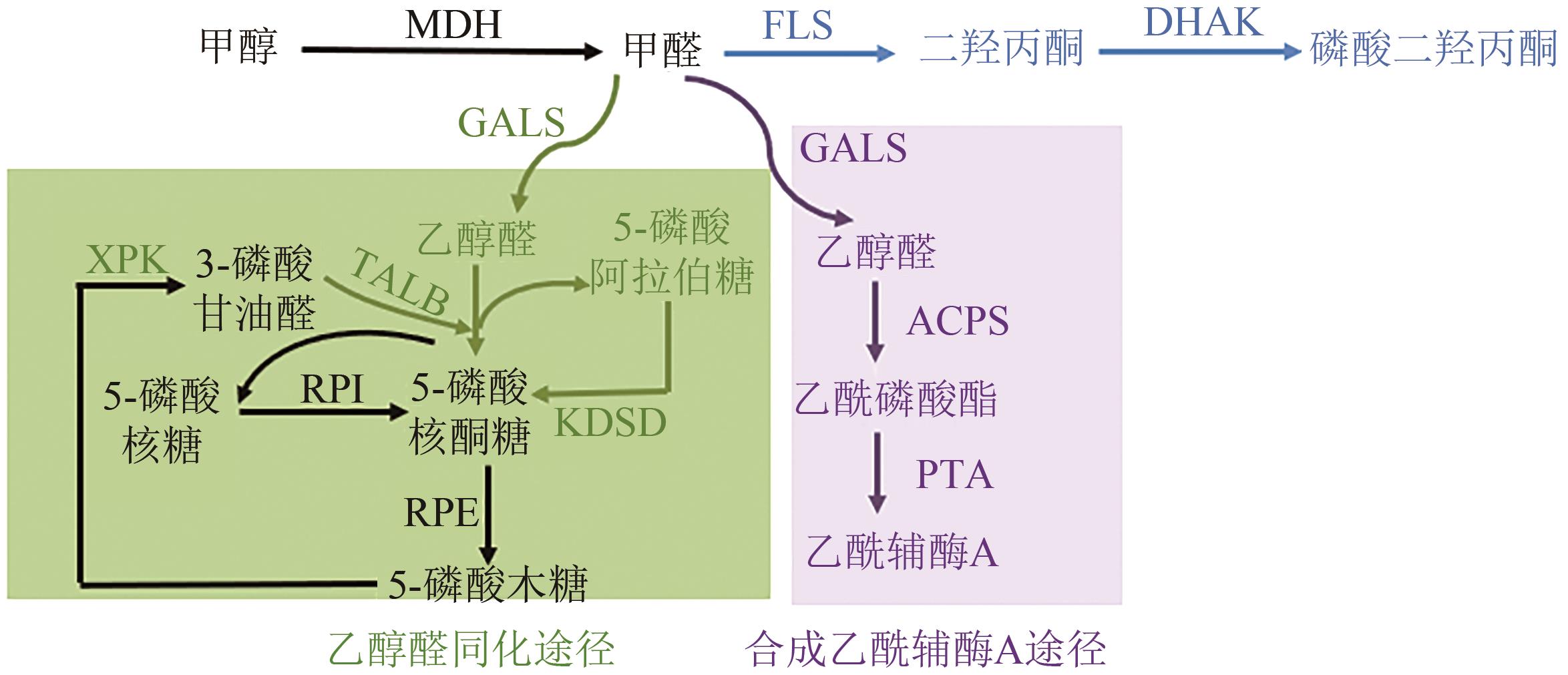
| 甲醛+甲醇+乙醇醛 | 体外到E. coli | 合成乙酰辅酶A(SACA)途径 | 热力学有利(ΔrG 乙醇醛的产率随甲醛底物浓度的增加而增加(2g·L-1时,转化率为80%) | [ |
| 甲醛 | 体外 | 体外构建醇醛同化(GAA)途径 | 碳转化率达到了88% | [ |
| 葡萄糖+甲醇 | E. coli Suc360 E. coli Suc460 E. coli Suc560 E. coli Suc660 | 引入甲醇芽孢杆菌的甲醇脱氢酶和不同供体生物的单磷酸核酮糖途径 | 丁二酸产率从(0.91±0.08)g·g-1提升至(0.98±0.11)g·g-1 | [ |
| 200mmol·L-1甲醇 | 无细胞体系 | 甲醇缩合循环(MCC) | 碳转化率为 80%(消耗33.5mm甲醇)(天然RuMP途径理论转化率为66%) | [ |
| 葡萄糖+甲酸盐 | E. coli SBS550MGCms243 | 将念珠菌依赖NAD+的甲酸脱氢酶基因(FDH1)与乳酸乳球菌丙酮酸羧化酶(PycA)共表达 | 改造后平均葡萄糖消耗速率为2g·L-1·h-1,琥珀酸生产强度为2g·L-1·h-1,副产物甲酸盐浓度为0~3mmol·L-1 | [ |
| 甲醇+葡萄糖 | E. coliΔfrmA | 共表达ACT蛋白提高MDH的活性 | MDH活性从22.1mU·mg-1 提升到29.1mU·mg-1 | [ |
| 底物 | 宿主 | 改造方法 | 改造结果 | 文献 |
|---|---|---|---|---|
| 甲酸盐 | fdoG MG1655-derived E. coli strain | 计算机设计甲醛酶(FLS)实现碳一到碳三化合物的合成 | 很高的化学推动力(>3kcal·mol-1,1cal=4.1840J) FLS活性增加了100倍 | [ |
| 甲醇+酵母膏 | BW25113frmA(EC-ΔfrmA) | 高效表达MDH和人工FLS 组装人工线性甲醇同化途径 | 甲醇消耗量从原始1g·L-1增加到1.45g·L-1 | [ |
| 甲醛+甲醇+乙醇醛 | 体外到E. coli | 合成乙酰辅酶A(SACA)途径 | 热力学有利(ΔrG 乙醇醛的产率随甲醛底物浓度的增加而增加(2g·L-1时,转化率为80%) | [ |
| 甲醛 | 体外 | 体外构建醇醛同化(GAA)途径 | 碳转化率达到了88% | [ |
| 葡萄糖+甲醇 | E. coli Suc360 E. coli Suc460 E. coli Suc560 E. coli Suc660 | 引入甲醇芽孢杆菌的甲醇脱氢酶和不同供体生物的单磷酸核酮糖途径 | 丁二酸产率从(0.91±0.08)g·g-1提升至(0.98±0.11)g·g-1 | [ |
| 200mmol·L-1甲醇 | 无细胞体系 | 甲醇缩合循环(MCC) | 碳转化率为 80%(消耗33.5mm甲醇)(天然RuMP途径理论转化率为66%) | [ |
| 葡萄糖+甲酸盐 | E. coli SBS550MGCms243 | 将念珠菌依赖NAD+的甲酸脱氢酶基因(FDH1)与乳酸乳球菌丙酮酸羧化酶(PycA)共表达 | 改造后平均葡萄糖消耗速率为2g·L-1·h-1,琥珀酸生产强度为2g·L-1·h-1,副产物甲酸盐浓度为0~3mmol·L-1 | [ |
| 甲醇+葡萄糖 | E. coliΔfrmA | 共表达ACT蛋白提高MDH的活性 | MDH活性从22.1mU·mg-1 提升到29.1mU·mg-1 | [ |
| 1 | OREN Yishai, MADELEINE Bouzon, Döring VOLKER, et al. In vivo assimilation of one-carbon via a synthetic reductive glycine pathway in Escherichia coli[J]. ACS Synthetic Biology, 2018, 7(9):2023-2028. |
| 2 | 高教琪, 周雍进. 甲醇生物转化的机遇与挑战[J]. 合成生物学, 2020, 1(2):158-173. |
| GAO Jiaoqi, ZHOU Yongjin. Advances in methanol bio-transformation[J]. Synthetic Biology Journal, 2020, 1(2):158-173. | |
| 3 | PRAMOTE Sirirote, TSUNEO Yamane, SHOICHI Shimizu. Production of L-serine from methanol and glycine by resting cells of a methylotroph under automatically controlled conditions[J]. Journal of Fermentation Technology, 1986, 64(5):89-396. |
| 4 | ANUPAMA, RAVINDRA P. Value-added food: single cell protein[J]. Biotechnology Advances, 2000, 18(6):459-479. |
| 5 | LIANG Weifan, CUI Lanyu, CUI Jinyu, et al. Biosensor-assisted transcriptional regulator engineering for Methylobacterium extorquens AM1 to improve mevalonate synthesis by increasing the acetyl-CoA supply[J]. Metabolic Engineering, 2017, 39:159-168. |
| 6 | ZHANG Xinying, WANG Denggang, DUAN Yehong, et al. Production of lycopene by metabolically engineered Pichia pastoris[J]. Bioscience Biotechnology and Biochemistry, 2020, 84(3):463-470. |
| 7 | BENNETT Robert Kyle, STEINBERG Lisa M, CHEN Wilfred, et al. Engineering the bioconversion of methane and methanol to fuels and chemicals in native and synthetic methylotrophs[J]. Current Opinion in Biotechnology, 2018, 50: 81-93. |
| 8 | ZHANG Wenming, ZHANG Ting, WU Sihua, et al. Guidance for engineering of synthetic methylotrophy based on methanol metabolism in methylotrophy[J]. RSC Advances, 2017, 7:4083-4091. |
| 9 | WHITAKER William Brain, SANDOVAL Nicholas, BWNNETT Robert Kyle, et al. Synthetic methylotrophy: engineering the production of biofuels and chemicals based on the biology of aerobic methanol utilization[J]. Current Opinion in Biotechnology, 2015, 33:165-175. |
| 10 | 陈阳, 杨雪, 袁倩倩, 等. 一碳化合物利用新途径的设计和体外构建[J]. 生物加工过程, 2017, 15(5): 86-92. |
| CHEN Yang, YANG Xue, YUAN Qianqian, et al. A computationally designed pathway for one carbon compounds utilization and its in vitro construction[J]. Chinese Journal of Bioprocess Engineering, 2017, 15(5): 86-92. | |
| 11 | MÜLLER Jonas E N, MEYERA Fabian, LITSANOV Boris. Engineering Escherichia coli for methanol conversion[J]. Metabolic Engineering, 2015, 28:190-201. |
| 12 | WHITAKER William Brain, JONESA Andrew, BWNNETT Robert Kyle, et al. Engineering the biological conversion of methanol to specialty chemicals in Escherichia coli[J]. Metabolic Engineering, 2017, 39:49-59. |
| 13 | WU Tungyun, CHEN Changting, LIU Jessica. Characterization and evolution of an activator-independent methanol dehydrogenase from Cupriavidus necator N-1[J]. Applied Microbiol Biotechnol, 2016, 100:4969-4983. |
| 14 | PRICE Vincent, CHEN Long, WHITAKER William Brain, et al. Scaffoldless engineered enzyme assembly for enhanced methanol utilization[J]. Proceedings of the National Academy of Sciences of the United States of America, 2016, 113(45): 12691-12696. |
| 15 | GONZALEZ Jacqueline, BWNNETT Robert Kyle, PAPOUTSAKIS Terry, et al. Methanol assimilation in Escherichia coli is improved by co-utilization of threonine and deletion of leucine-responsive regulatory protein[J]. Metabolic Engineering, 2018, 45:67-74. |
| 16 | MEYER Fabian, KELLER Philipp, HARTL Johannes, et al. Methanol-essential growth of Escherichia coli[J]. Nature Communications, 2018, 9:1508. |
| 17 | HE Hai, CHRISTIAN Edlich Muth, STEFFEN Lindner. Ribulose monophosphate shunt provides nearly all biomass and energy required for growth of E. coli[J]. American Chemical Society Synthetic Biology, 2018, 7(6):1601-1611. |
| 18 | BWNNETT Robert Kyle, GONZALEZ Jacqueline, WHITAKER William Brain. Expression of heterologous non-oxidative pentose phosphate pathway from Bacillus methanolicus and phosphoglucose isomerase deletion improves methanol assimilation and metabolite production by a synthetic Escherichia coli methylotroph[J]. Metabolic Engineering, 2018, 45: 75-85. |
| 19 | WOOLSTON Benjamin, KING Jason, REITER Michael, et al. Improving formaldehyde consumption drivesmethanol assimilation in engineered E. coli[J]. Nature Communications, 2018, 9(1): 1-12. |
| 20 | ZHANG Wenming, SONG Meng, YANG Qiao, et al. Current advance in bioconversion of methanol to chemicals[J]. Biotechnology for Biofuels, 2018, 11: 260. |
| 21 | CHEN Y H, HSIN-WEI J, TSUEI C Y, et al. Converting Escherichia coli to a synthetic methylotroph growing solely on methanol[J]. Cell, 2020, 4(182): 933-946. |
| 22 | BENNETT Robert Kyle, DILLON Michael, AGEE Alec, et al. Engineering Escherichia coli for methanol-dependent growth on glucose for metabolite production[J]. Metabolic Engineering, 2020, 60:45-55. |
| 23 | CHEN C T, WU T Y, CHEN F, et al. Synthetic methanol auxotrophy of Escherichia coli for methanol-dependent growth and production[J]. Metabolic Engineering, 2018, 49: 257-266. |
| 24 | YU Hong, LIAO James. A modified serine cycle in Escherichia coli coverts methanol and CO2 to two-carbon compounds[J]. Nature Communications, 2018, 9: 3992. |
| 25 | KALYUZHNAYA Marina, LIDSTROM Mary. QscR-mediated transcriptional activation of serine cycle genes in methylobacterium extorquens AM1[J]. Journal of Bacteriology, 2005, 21(187): 7511-7517. |
| 26 | KALYUZHNAYA Marina, LIDSTROM Mary. QscR, a LysR-type transcriptional regulator and CbbR Homolog, is involved in regulation of the serine cycle genes in Methylobacterium extorquens AM1 [J]. Journal of Bacteriology, 2003, 4(185): 1229-1235. |
| 27 | OREN Yishai, LEANDER Goldbach, HEZI Tenenboim, et al. Engineered assimilation of exogenous and endogenous formate in Escherichia coli[J]. ACS Synthetic Biology, 2017, 6: 1722-1731. |
| 28 | HE Hai, Höper RUNE, MORITZ Dodenhöft, et al. An optimized methanol assimilation pathway relying on promiscuous formaldehyde-condensing aldolases in E.coli[J]. Metabolic Engineering, 2020, 60: 1-13. |
| 29 | WANG Xiaolu, WANG Yu, LIU Jiao, et al. Biological conversion of methanol by evolved Escherichia coli carrying a linear methanol assimilation pathway[J]. Bioresources and Bioprocessing, 2017, 4: 41. |
| 30 | LU Xiaoyun, LIU Yuwan, YANG Yiqun, et al. Constructing a synthetic pathway for acetyl-coenzyme a from one-carbon through enzyme design[J]. Nature Communications, 2019, 10(1): 1387. |
| 31 | YANG Xue, YUAN Qianqian, LUO Hao, et al. Systematic design and in vitro validation of novel one-carbon assimilation pathways[J]. Metabolic Engineering, 2019, 56:142-153. |
| 32 | LIAO James, MI Luo, PONTRELLI Sammy, et al. Fuelling the future: microbial engineering for the production of sustainable biofuels[J]. Nature Reviews Microbiology, 2016, 14(5):288-304. |
| 33 | SIEGEL Justin, SMITH Amada Lee, POUST Sean, et al. Computational protein design enables a novel one-carbon assimilation pathway[J]. Proceedings of the National Academy of Sciences of the United States of America, 2015, 112(12):3704-3709. |
| 34 | ZHANG Wenming, ZHANG Ting, SONG Meng, et al. Metabolic engineering of Escherichia coli for high yield production of succinic acid driven by methanol[J]. ACS Synthetic Biology, 2018, 7(12):2803-2811. |
| 35 | BOGORAD Igor, CHENA Changting, THEISENA Matthew, et al. Building carbon-carbon bonds using a biocatalytic methanol condensation cycle[J]. Proceedings of the National Academy of Sciences of the United States of America, 2014, 45(111):15928-15933. |
| 36 | BALZER Grant, THAKKER Chandresh, BENNETT George, et al. Metabolic engineering of Escherichia coli to minimize byproduct formate and improving succinate productivity through increasing NADH availability by heterologous expression of NAD+-dependent formate dehydrogenase[J]. Metabolic Engineering, 2013, 20:1-8. |
| 37 | WANG Xin, WANG Xuelin, LU Xiaolu, et al. Methanol fermentation increases the production of NAD(P)H-dependent chemicals in synthetic methylotrophic Escherichia coli[J]. Biotechnology for Biofuels, 2019, 12:17. |
| [1] | SHI Yongxing, LIN Gang, SUN Xiaohang, JIANG Weigeng, QIAO Dawei, YAN Binhang. Research progress on active sites in Cu-based catalysts for CO2 hydrogenation to methanol [J]. Chemical Industry and Engineering Progress, 2023, 42(S1): 287-298. |
| [2] | XU Jiaheng, LI Yongsheng, LUO Chunhuan, SU Qingquan. Optimization of methanol steam reforming process [J]. Chemical Industry and Engineering Progress, 2023, 42(S1): 41-46. |
| [3] | SHU Bin, CHEN Jianhong, XIONG Jian, WU Qirong, YU Jiangtao, YANG Ping. Necessity analysis of promoting the development of green methanol under the goal of carbon neutrality [J]. Chemical Industry and Engineering Progress, 2023, 42(9): 4471-4478. |
| [4] | WANG Zijian, KE Ming, SONG Zhaozheng, LI Jiahan, TONG Yanbing, SUN Jinru. Progress in alkylation of gasoline with molecular sieve catalyst for benzene reduction [J]. Chemical Industry and Engineering Progress, 2023, 42(5): 2371-2389. |
| [5] | XIAO Yaoxin, ZHANG Jun, HU Sheng, SHAN Rui, YUAN Haoran, CHEN Yong. Cu-Zn catalyzed hydrogenation of furfural with methanol as hydrogen donor [J]. Chemical Industry and Engineering Progress, 2023, 42(3): 1341-1352. |
| [6] | HUANG Qizhong, LIU Bing, MA Hongpeng, LYU Wenjie. Methanol to olefin wastewater treatment based on a novel microchannel separation technology [J]. Chemical Industry and Engineering Progress, 2023, 42(2): 669-676. |
| [7] | GUO Feng, ZHANG Shangjie, JIANG Yujia, JIANG Wankui, XIN Fengxue, ZHANG Wenming, JIANG Min. Biotransformation of one-carbon resources by yeast [J]. Chemical Industry and Engineering Progress, 2023, 42(1): 30-39. |
| [8] | ZHAO Tongxin, ZHAO Lei, ZHANG Yanping, LI Yin. Research progress of formic acid biotransformation [J]. Chemical Industry and Engineering Progress, 2023, 42(1): 67-72. |
| [9] | LIU Penglong, XU Xiongfei, ZHANG Wei, XU Xin, ZHANG Kan, WANG Junwen. Local modeling and optimization of K-means-PSO-SVR for methanol to aromatics [J]. Chemical Industry and Engineering Progress, 2022, 41(9): 4691-4700. |
| [10] | ZHANG Jiaqi, LIN Lina, GAO Wengui, ZHU Xing. Effect of CeO2 morphology on the performance of CuO/CeO2 catalyst for CO2 hydrogenation to methanol [J]. Chemical Industry and Engineering Progress, 2022, 41(8): 4213-4223. |
| [11] | XIANG Sheng, WANG Chao, ZHUANG Yu, GU Siwen, ZHANG Lei, DU Jian. Design and control of pressure-swing distillation for separating methyl acetate-methanol-ethyl acetate azeotropic system [J]. Chemical Industry and Engineering Progress, 2022, 41(8): 4065-4076. |
| [12] | ZHAO Jianbing, YANG Dan, SHU Yuancao, ZHU Junbo, PU Shiping, SONG Xiaodan, LIU Shouqing, CHAI Xijuan, LI Xuemei. Preparation of Na2CO3 /CF solid base and its catalytic transesterification of rapeseed oil [J]. Chemical Industry and Engineering Progress, 2022, 41(7): 3608-3614. |
| [13] | LI Guixian, ZHANG Junqiang, YANG Yong, FAN Xueying, WANG Dongliang. A novel PX production shortcut through PX selectivity intensification in toluene and methanol methylation [J]. Chemical Industry and Engineering Progress, 2022, 41(6): 2939-2947. |
| [14] | WANG Jijie, HAN Zhe, CHEN Siyu, TANG Chizhou, SHA Feng, TANG Shan, YAO Tingting, LI Can. Liquid sunshine methanol [J]. Chemical Industry and Engineering Progress, 2022, 41(3): 1309-1317. |
| [15] | CHANG Fei, ZHAN Guoxiong, SHI Sensen, ZENG Shaojuan, ZHANG Xiangping. Process assessment for electroreduction CO2 to methanol in ionic liquid electrolyte [J]. Chemical Industry and Engineering Progress, 2022, 41(3): 1256-1264. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||