化工进展 ›› 2019, Vol. 38 ›› Issue (01): 598-605.DOI: 10.16085/j.issn.1000-6613.2018-1138
酿酒酵母高效合成萜类化合物的组合调控策略
- 北京理工大学化学与化工学院,北京 100081
-
收稿日期:2018-05-31修回日期:2018-07-01出版日期:2019-01-05发布日期:2019-01-05 -
通讯作者:李春 -
作者简介:常鹏程(1993—),男,硕士研究生,研究方向为代谢工程与合成生物学。E-mail:<email>changpengcheng301@163.com</email>。|李春,教授,博士生导师,研究方向生物转化与酶工程、代谢工程与合成生物学。E-mail:<email>lichun@bit.edu.cn</email>。 -
基金资助:国家杰出青年科学基金(21425624);国家自然科学基金(21606018);国家杰出青年科学基金(21425624);国家自然科学基金(21606018)。
Combinatorial regulation strategies for efficient synthesis of terpenoids in Saccharomyces cerevisiae
Pengcheng CHANG( ),Yang YU,Ying WANG,Chun LI(
),Yang YU,Ying WANG,Chun LI( )
)
- School of Chemistry and Chemical Engineering,Beijing Institute of Technology,Beijing 100081
-
Received:2018-05-31Revised:2018-07-01Online:2019-01-05Published:2019-01-05 -
Contact:Chun LI
摘要:
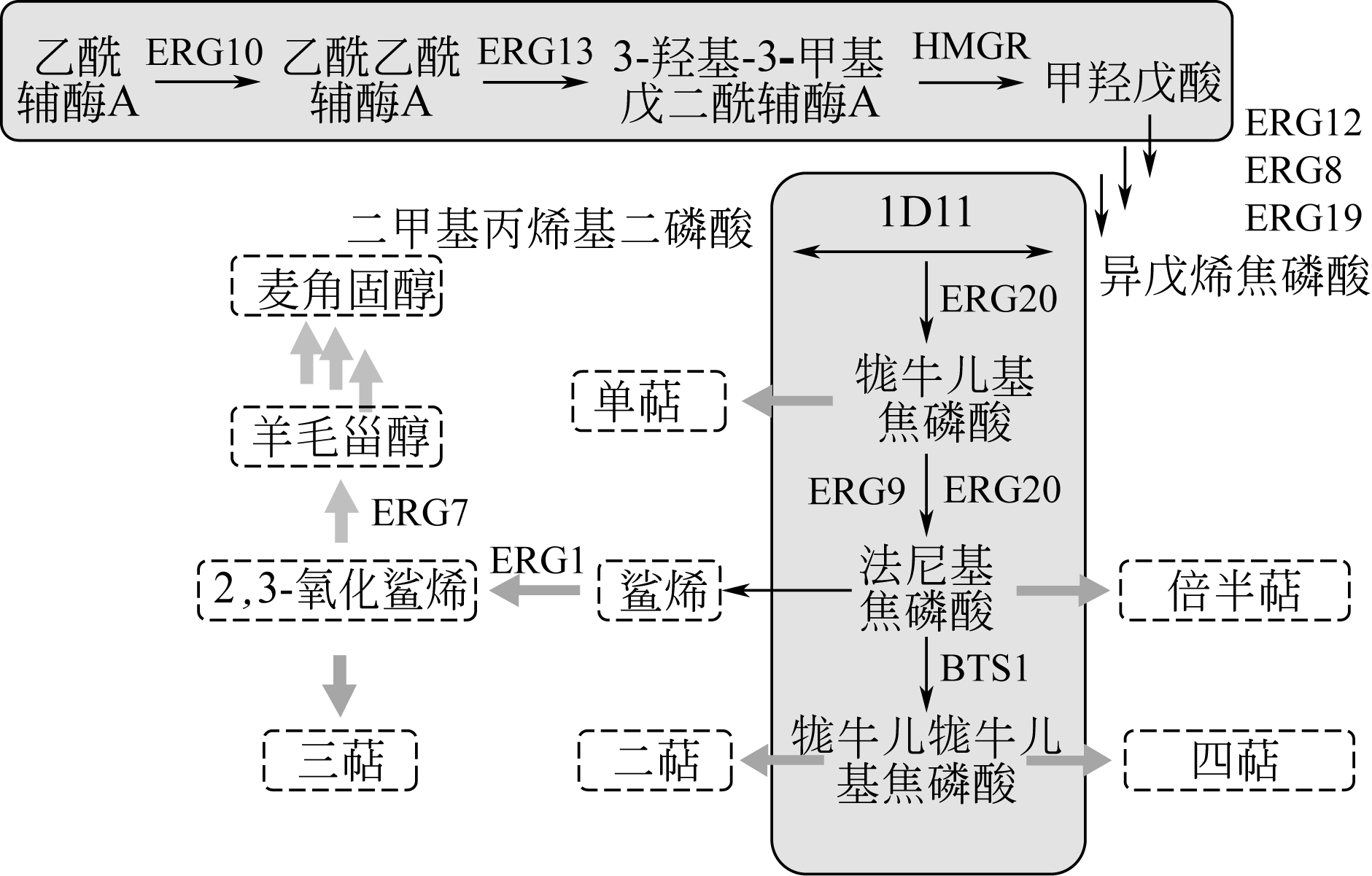
萜类化合物具有广泛的生理活性与重要的经济价值,利用酿酒酵母进行萜类合成具有低价、高效等优势。然而部分植物源合成萜类的关键酶在酿酒酵母中难表达、产量低,难以工业应用,因此有效的调控策略显得至关重要。本文从萜类化合物在酿酒酵母中的合成途径入手,介绍了关键酶、代谢途径、CRISPR基因编辑系统和人工合成染色体技术4个方面的调控策略在酿酒酵母合成萜类化合物中的应用。阐述了关键酶的筛选、改造,理性与非理性设计,MVA途径、乙酰辅酶A合成途径与亚细胞结构的代谢途径改造的优势。指出了多重调控策略组合调控的方式是实现酿酒酵母高效合成萜类化合物的有效方法。此外,CRISPR基因编辑系统与人工合成染色体技术的快速发展将为酿酒酵母细胞工厂的深入开发与利用提供有力工具。
中图分类号:
引用本文
常鹏程, 于洋, 王颖, 李春. 酿酒酵母高效合成萜类化合物的组合调控策略[J]. 化工进展, 2019, 38(01): 598-605.
Pengcheng CHANG, Yang YU, Ying WANG, Chun LI. Combinatorial regulation strategies for efficient synthesis of terpenoids in Saccharomyces cerevisiae[J]. Chemical Industry and Engineering Progress, 2019, 38(01): 598-605.
| 萜类 | 合成量 | 萜类 | 合成量 |
|---|---|---|---|
| 牻牛儿醇 | 36.04[ | 铁锈醇 | 10[ |
| 椒二醇 | 50[ | β-香树脂醇 | 138.80[ |
| 紫衫二烯 | 8.7[ | 甘草次酸 | 18.9[ |
| 次丹参酮二烯 | 488mg/L[ | 齐敦果酸 | 606.9mg/L[ |
表1 部分酿酒酵母合成萜类的产量
| 萜类 | 合成量 | 萜类 | 合成量 |
|---|---|---|---|
| 牻牛儿醇 | 36.04[ | 铁锈醇 | 10[ |
| 椒二醇 | 50[ | β-香树脂醇 | 138.80[ |
| 紫衫二烯 | 8.7[ | 甘草次酸 | 18.9[ |
| 次丹参酮二烯 | 488mg/L[ | 齐敦果酸 | 606.9mg/L[ |
| 1 | PEMBERTON T A , CHEN M B , HARRIS G G , et al .Exploring the influence of domain architecture on the catalytic function of diterpene synthases[J].Biochemistry, 2017, 56(14): 2010-2023. |
| 2 | VANEGAS K G , LEHKA B J , MORTENSEN U H .SWITCH:a dynamic CRISPR tool for genome engineering and metabolic pathway control for cell factory construction in Saccharomyces cerevisiae [J].Microbial Cell Factories, 2017, 16(1): 25. |
| 3 | ALPER H , MIYAOKU K , STEPHANOPOULOS G .Construction of lycopene-overproducing E. coli strains by combining systematic and combinatorial gene knockout targets[J].Nature Biotechnology, 2005, 23(5): 612-616. |
| 4 | ZHAO Y J , FAN J J , WANG C , et al . Enhancing oleanolic acid production in engineered Saccharomyces cerevisiae [J].Bioresource Technology, 2018, 257:339-343. |
| 5 | KIM K A , LEE J S , PARK H J , et al .Inhibition of cytochrome P450 activities by oleanolic acid and ursolic acid in human liver microsomes[J].Life Sciences, 2004, 74(22): 2769-2779. |
| 6 | 朱明, 王彩霞, 李春 . 工程化酿酒酵母合成植物三萜类化合物[J]. 化工学报, 2015, 66(9): 3350-3356. |
| ZHU M , WANG C X , LI C . Engineered Saccharomyces cerevisiae for biosynthesis of plant triterpenoids[J]. CIESC Journal, 2015, 66(9): 3350-3356. | |
| 7 | CHAN W K , TAN L T H , CHAN K G , et al . Nerolidol: a sesquiterpene alcohol with multi-faceted pharmacological and biological activities[J]. Molecules, 2016, 21(5): 529. |
| 8 | HU G Y , PENG C , XIE X F , et al . Availability, pharmaceutics, security, pharmacokinetics, and pharmacological activities of patchouli alcohol[J]. Evidence-Based Complementary and Alternative Medicine, 2017(4): 1-9. |
| 9 | PADDON C J , WESTFALL P J , PITERA D J , et al . High-level semi-synthetic production of the potent antimalarial artemisinin[J]. Nature, 2013, 496(7446): 528-532. |
| 10 | LIU J D , ZHANG W P , DU G C , et al . Overproduction of geraniol by enhanced precursor supply in Saccharomyces cerevisiae [J]. Journal of Biotechnology, 2013, 168(4): 446-451. |
| 11 | TAKAHASHI S , YEO Y , GREENHAGEN B T , et al . Metabolic engineering of sesquiterpene metabolism in yeast[J]. Biotechnology and Bioengineering, 2007, 97(1): 170-181. |
| 12 | ENGELS B , DAHM P , JENNEWEIN S . Metabolic engineering of taxadiene biosynthesis in yeast as a first step towards Taxol (Paclitaxel) production[J]. Metabolic Engineering, 2008, 10(3/4): 201-206. |
| 13 | DAI Z B , LIU Y , HUANG L Q , et al . Production of miltiradiene by metabolically engineered Saccharomyces cerevisiae [J]. Biotechnology and Bioengineering, 2012, 109(11): 2845–2853. |
| 14 | GUO J , ZHOU Y J , HILLWIG M L , et al . CYP76AH1 catalyzes turnover of miltiradiene in tanshinones biosynthesis and enables heterologous production of ferruginol in yeasts[J]. Proceedings of the National Academy of Sciences, 2013, 110(29): 12108-12113. |
| 15 | ZHANG G L , CAO Q , LIU J Z , et al . Refactoring β-amyrin synthesis in Saccharomyces cerevisiae [J]. AIChE Journal, 2015, 61(10): 3172-3179. |
| 16 | ZHU M , WANG C X , SUN W T , et al . Boosting 11-oxo-β-amyrin and glycyrrhetinic acid synthesis in Saccharomyces cerevisiae via pairing novel oxidation and reduction system from legume plants[J]. Metabolic Engineering, 2018, 45:43-50. |
| 17 | CHEN Y , XIAO W H , WANG Y , et al . Lycopene overproduction in Saccharomyces cerevisiae through combining pathway engineering with host engineering[J]. Microbial Cell Factories, 2016, 15(1): 113. |
| 18 | BOHLMANN J , MEYER-GAUEN G , CROTEAU R . Plant terpenoid synthases: molecular biology and phylogenetic analysis[J]. Proceedings of the National Academy of Sciences, 1998, 95(8): 4126-4133. |
| 19 | ZHAO J Z , BAO X M , LI C , et al . Improving monoterpene geraniol production through geranyl diphosphate synthesis regulation in Saccharomyces cerevisiae [J]. Applied Microbiology and Biotechnology, 2016, 100(10): 4561-4571. |
| 20 | JIANG G Z , YAO M D , WANG Y , et al . Manipulation of GES and ERG20 for geraniol overproduction in Saccharomyces cerevisiae [J]. Metabolic Engineering, 2017, 41:57-66. |
| 21 | SCHMIDT-DANNERT C , ARNOLD F H . Directed evolution of industrial enzymes[J]. Trends in Biotechnology, 1999, 17(4): 135-136. |
| 22 | XIE W P , LV X M , YE L D , et al . Construction of lycopene-overproducing Saccharomyces cerevisiae by combining directed evolution and metabolic engineering[J]. Metabolic Engineering, 2015, 30:69-78. |
| 23 | WANG F , LV X M , XIE W P , et al . Combining Gal4p-mediated expression enhancement and directed evolution of isoprene synthase to improve isoprene production in Saccharomyces cerevisiae [J]. Metabolic Engineering, 2016, 39:257-266. |
| 24 | ZHUANG Y , YANG G Y , CHEN X H , et al . Biosynthesis of plant-derived ginsenoside Rh2 in yeast via repurposing a key promiscuous microbial enzyme[J]. Metabolic Engineering, 2017, 42:25-32. |
| 25 | DING M Z , YAN H F , LI L F , et al . Biosynthesis of taxadiene in Saccharomyces cerevisiae:selection of geranylgeranyl diphosphate synthase directed by a computer-aided docking strategy[J]. PLoS One, 2014, 9(10): e109348. |
| 26 | LISCUM L , FINERMOORE J , STROUD R M , et al . Domain structure of 3-hydroxy-3-methylglutaryl coenzyme A reductase, a glycoprotein of the endoplasmic reticulum[J]. Journal of Biological Chemistry, 1985, 260(1): 522-530. |
| 27 | BASSON M E , THORSNESS M , FINER-MOORE J , et al . Structural and functional conservation between yeast and human 3-hydroxy-3-methylglutaryl coenzyme A reductases, the rate-limiting enzyme of sterol biosynthesis[J]. Molecular and Cellular Biology., 1988, 8(9): 3797-3808. |
| 28 | POLAKOWSKI T , STAHL U , LANG C . Overexpression of a cytosolic hydroxymethylglutaryl-CoA reductase leads to squalene accumulation in yeast [J]. Applied Microbiology and Biotechnology, 1998, 49(1): 66-71. |
| 29 | KEASLING J D . Synthetic biology and the development of tools for metabolic engineering[J]. Metabolic Engineering, 2012, 14(3): 189-195. |
| 30 | MANTZOURIDOU F , TSIMIDOU M Z . Observations on squalene accumulation in Saccharomyces cerevisiae due to the manipulation of HMG2 and ERG6[J]. FEMS Yeast Research, 2010, 10(6): 699-707. |
| 31 | LIAN J Z , SI T , NAIR N U , et al . Design and construction of acetyl-CoA overproducing Saccharomyces cerevisiae strains[J]. Metabolic Engineering, 2014, 24(7): 139-149. |
| 32 | CHEN Y , DAVIET L , SCHALK M , et al . Establishing a platform cell factory through engineering of yeast acetyl-CoA metabolism[J]. Metabolic Engineering, 2013, 15(1): 48-54. |
| 33 | FARHI M , MARHEVKA E , MASCI T , et al . Harnessing yeast subcellular compartments for the production of plant terpenoids[J]. Metabolic Engineering, 2011, 13(5): 474-481. |
| 34 | YUAN J F , CHING C B . Mitochondrial acetyl-CoA utilization pathway for terpenoid productions[J]. Metabolic Engineering, 2016, 38:303-309. |
| 35 | LV X M , WANG F , ZHOU P P , et al . Dual regulation of cytoplasmic and mitochondrial acetyl-CoA utilization for improved isoprene production in Saccharomyces cerevisiae [J]. Nature Communications, 2016, 7:12851. |
| 36 | ARENDT P , MIETTINEN K , POLLIER J , et al . An endoplasmic reticulum-engineered yeast platform for overproduction of triterpenoids[J]. Metabolic Engineering, 2017, 40:165-175. |
| 37 | ASADOLLAHI M A , MAURY J , MøLLER K , et al . Production of plant sesquiterpenes in Saccharomyces cerevisiae:effect of ERG9 repression on sesquiterpene biosynthesis[J]. Biotechnology and Bioengineering, 2008, 99(3): 666-677. |
| 38 | SCALCINATI G , PARTOW S , SIEWERS V , et al . Combined metabolic engineering of precursor and co-factor supply to increase α santalene production by Saccharomyces cerevisiae [J]. Microbial Cell Factories, 2012, 11(1): 117. |
| 39 | PENG B Y , PLAN M R , CHRYSANTHOPOULOS P , et al . A squalene synthase protein degradation method for improved sesquiterpene production in Saccharomyces cerevisiae [J]. Metabolic Engineering, 2016, 39:209-219. |
| 40 | CONG L , RAN F A , COX D , et al . Multiplex genome engineering using CRISPR/Cas systems[J]. Science, 2013, 339(6121): 819-823. |
| 41 | JAKOČIŪNAS T , BONDE I , HERRGåRD M , et al . Multiplex metabolic pathway engineering using CRISPR/Cas9 in Saccharomyces cerevisiae [J]. Metabolic Engineering, 2015, 28:213-222. |
| 42 | SHI S B , LIANG Y Y , ZHANG M M , et al . A highly efficient single-step, markerless strategy for multi-copy chromosomal integration of large biochemical pathways in Saccharomyces cerevisiae [J]. Metabolic Engineering, 2016, 33:19-27. |
| 43 | BARBIERI E M , MUIR P , AKHUETIE-ONI B O , et al . Precise editing at DNA replication forks enables multiplex genome engineering in eukaryotes [J]. Cell, 2017, 171(6): 1453-1467. |
| 44 | ZALATAN J G , LEE M E , ALMEIDA R , et al . Engineering complex synthetic transcriptional programs with CRISPR RNA scaffolds[J]. Cell, 2015, 160(1/2): 339-350. |
| 45 | GILBERT L A , LARSON M H , MORSUT L , et al . CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes[J]. Cell, 2013, 154(2): 442-451. |
| 46 | LIAN J Z , HAMEDIRAD M , HU S , et al . Combinatorial metabolic engineering using an orthogonal tri-functional CRISPR system[J]. Nature Communications, 2017, 8(1): 1688. |
| 47 | DYMOND J S , RICHARDSON S M , COOMBES C E , et al . Synthetic chromosome arms function in yeast and generate phenotypic diversity by design [J]. Nature, 2011, 477(7365): 471-476. |
| 48 | SHEN Y , WANG Y , CHEN T , et al . Deep functional analysis of synII, a770 kb synthetic yeast chromosome[J]. Science, 2017, 355(6329): eaaf4791. |
| 49 | XIE Z X , LI B Z , MITCHELL L A , et al . "Perfect" designer chromosome V and behavior of a ring derivative[J]. Science, 2017, 355(6329): eaaf4704. |
| 50 | MITCHELL L A , WANG A , STRACQUADANIO G , et al . Synthesis, debugging, and effects of synthetic chromosome consolidation:synⅥ and beyond[J]. Science, 2017, 355(6329): eaaf4831. |
| 51 | WU Y , LI B Z , ZHAO M , et al . Bug mapping and fitness testing of chemically synthesized chromosome X[J]. Science, 2017, 355(6329): eaaf4706. |
| 52 | ZHANG W M , ZHAO G H , LUO Z Q , et al . Engineering the ribosomal DNA in a megabase synthetic chromosome[J]. Science, 2017, 355(6329): eaaf3981. |
| 53 | ANNALURU N , MULLER H , MITCHELL L A , et al . Total synthesis of a functional designer eukaryotic chromosome[J]. Science, 2014, 344(6179): 55-58. |
| 54 | WU Y , ZHU R Y , MITCHELL L A , et al . In vitro DNA SCRaMbLE[J]. Nature Communications, 2018, 9(1): 1935. |
| 55 | SHEN M J , WU Y , YANG K , et al . Heterozygous diploid and interspecies SCRaMbLEing[J]. Nature Communications, 2018, 9(1): 1934. |
| 56 | LIU W , LUO Z Q , WANG Y , et al . Rapid pathway prototyping and engineering using in vitro and in vivo synthetic genome SCRaMbLE-in methods [J]. Nature Communications, 2018, 9(1): 1936. |
| 57 | JIA B , WU Y , LI B Z , et al . Precise control of SCRaMbLE in synthetic haploid and diploid yeast[J]. Nature Communications, 2018, 9(1): 1933. |
| [1] | 陶雨萱, 郭亮, 高聪, 宋伟, 陈修来. 代谢工程改造微生物固定二氧化碳研究进展[J]. 化工进展, 2023, 42(1): 40-52. |
| [2] | 郭峰, 张尚杰, 蒋羽佳, 姜万奎, 信丰学, 章文明, 姜岷. 一碳资源在酵母中的利用与转化[J]. 化工进展, 2023, 42(1): 30-39. |
| [3] | 董晓宇. 酿酒酵母钙通道膜蛋白单克隆抗体制备及鉴定[J]. 化工进展, 2021, 40(S1): 334-343. |
| [4] | 陶雨萱, 张尚杰, 景艺文, 信丰学, 董维亮, 周杰, 蒋羽佳, 章文明, 姜岷. 甲基营养型大肠杆菌构建策略的研究进展[J]. 化工进展, 2021, 40(7): 3932-3941. |
| [5] | 李玲, 于泳, 胡永红. 发酵法生产利普司他汀的研究进展[J]. 化工进展, 2021, 40(4): 2251-2257. |
| [6] | 郭亮, 高聪, 张丽, 陈修来, 刘立明. 人工代谢路径适配性的研究进展[J]. 化工进展, 2021, 40(3): 1252-1261. |
| [7] | 王颖, 曲俊泽, 梁楠, 郝鹤, 元英进. 合成类胡萝卜素细胞工厂的快速构建和定向进化[J]. 化工进展, 2021, 40(3): 1187-1201. |
| [8] | 周子康, 许平. 全局转录调控在细胞工厂构建中的应用与进展[J]. 化工进展, 2021, 40(3): 1248-1251. |
| [9] | 刘卫兵, 叶邦策. 放线菌聚酮类化合物的合成生物学研究及生物制造[J]. 化工进展, 2021, 40(3): 1226-1237. |
| [10] | 孙文涛, 李春. 微生物合成植物天然产物的细胞工厂设计与构建[J]. 化工进展, 2021, 40(3): 1202-1214. |
| [11] | 马悦原, 陈金春, 陈国强. 嗜盐微生物底盘细胞:应用和前景[J]. 化工进展, 2021, 40(3): 1178-1186. |
| [12] | 高聪, 郭亮, 胡贵鹏, 陈修来, 刘立明. 微生物细胞工厂碳流调控进展[J]. 化工进展, 2021, 40(12): 6807-6817. |
| [13] | 王琛, 赵猛, 丁明珠, 王颖, 姚明东, 肖文海. 生物支架系统在合成生物学中的应用[J]. 化工进展, 2020, 39(11): 4557-4567. |
| [14] | 陈露,刘丁玉,汪保卫,赵玉姣,贾广韬,陈涛,王智文. 大肠杆菌乙酰辅酶A代谢调控及其应用研究进展[J]. 化工进展, 2019, 38(9): 4218-4226. |
| [15] | 程申, 张颂红, 贠军贤. α-酮异己酸的生物合成研究进展[J]. 化工进展, 2018, 37(12): 4821-4829. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||