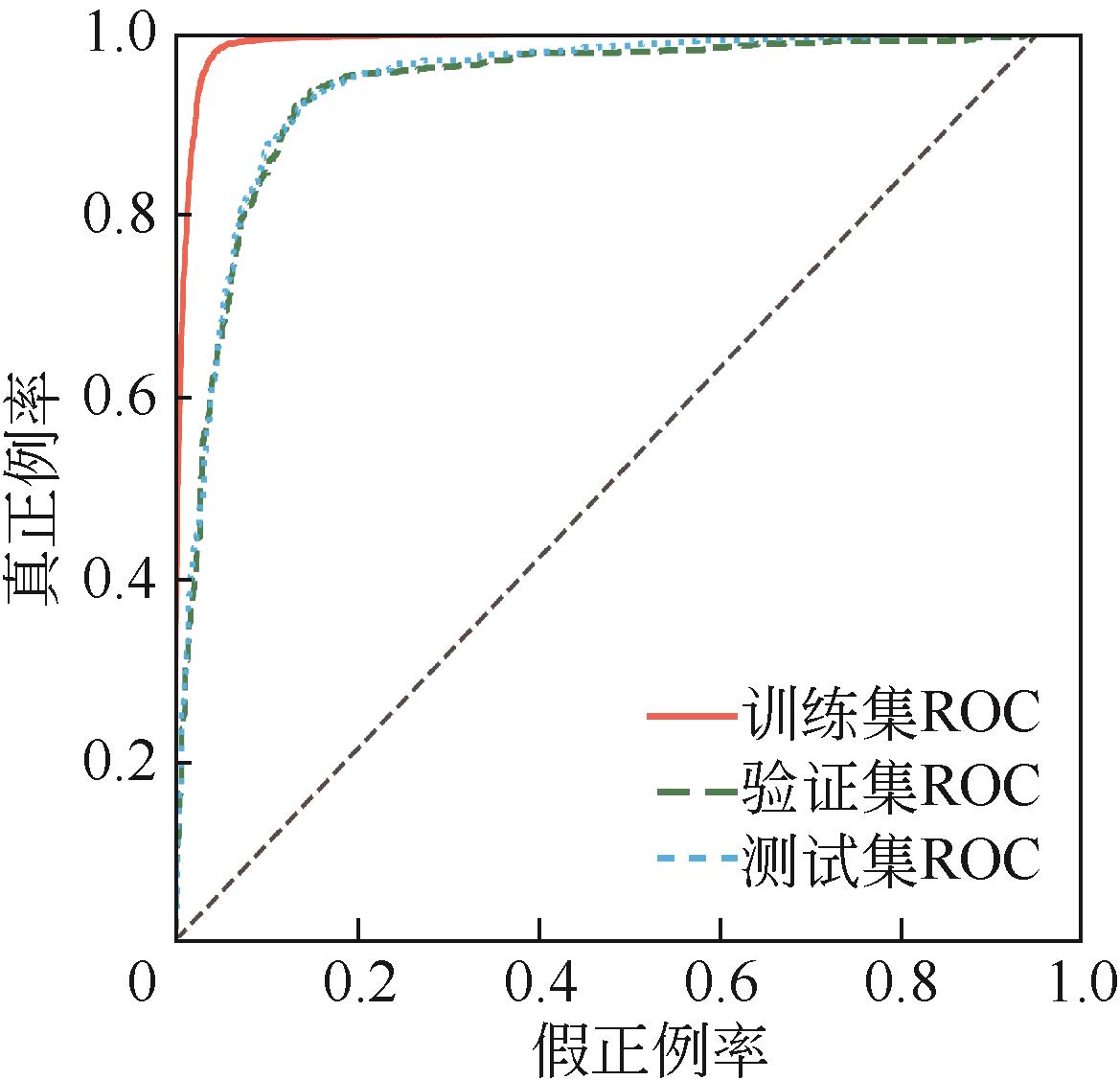
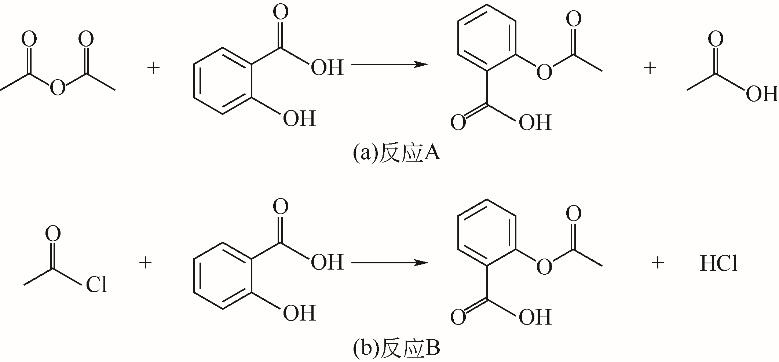
| 1 |
LI Baiqing, CHEN Hongming. Prediction of compound synthesis accessibility based on reaction knowledge graph[J]. Molecules, 2022, 27(3): 1039.
|
| 2 |
QIU Fayang. Strategic efficiency—The new thrust for synthetic organic chemists[J]. Canadian Journal of Chemistry, 2008, 86(9): 903-906.
|
| 3 |
Grzegorz SKORACZYŃSKI, KITLAS Mateusz, Błażej MIASOJEDOW, et al. Critical assessment of synthetic accessibility scores in computer-assisted synthesis planning[J]. Journal of Cheminformatics, 2023, 15(1): 1-9.
|
| 4 |
Milan VORŠILÁK, Michal KOLÁŘ, Ivan ČMELO, et al. SYBA: Bayesian estimation of synthetic accessibility of organic compounds[J]. Journal of Cheminformatics, 2020, 12(1): 1-13.
|
| 5 |
ITO Shun, BABA Yukino, ISOMURA Tetsu, et al. Synthetic accessibility assessment using auxiliary responses[J]. Expert Systems With Applications, 2020, 145: 113106.
|
| 6 |
COLEY C W, ROGERS L, GREEN W H, et al. SCScore: Synthetic complexity learned from a reaction corpus[J]. Journal of Chemical Information and Modeling, 2018, 58(2): 252-261.
|
| 7 |
YU Jiahui, WANG Jike, ZHAO Hong, et al. Organic compound synthetic accessibility prediction based on the graph attention mechanism[J]. Journal of Chemical Information and Modeling, 2022, 62(12): 2973-2986.
|
| 8 |
ERTL Peter, SCHUFFENHAUER Ansgar. Estimation of synthetic accessibility score of drug-like molecules based on molecular complexity and fragment contributions[J]. Journal of Cheminformatics, 2009, 1(1): 8.
|
| 9 |
ZHAO Yanan, LIU Xiaochen, LU Han, et al. An optimized deep convolutional neural network for yield prediction of Buchwald-Hartwig amination[J]. Chemical Physics, 2021, 550: 111296.
|
| 10 |
THAKKAR A, CHADIMOVÁ V, BJERRUM E J, et al. Retrosynthetic accessibility score (RAscore)–rapid machine learned synthesizability classification from AI driven retrosynthetic planning[J]. Chemical Science, 2021, 12(9): 3339-3349.
|
| 11 |
SHUI Zeren, KARYPIS George. Heterogeneous molecular graph neural networks for predicting molecule properties[C]//2020 IEEE International Conference on Data Mining (ICDM). November 17-20, 2020, Sorrento, Italy. IEEE, 2021: 492-500.
|
| 12 |
YOU Jiaxuan, LIU Bowen, YING Rex, et al. Graph convolutional policy network for goal-directed molecular graph generation[J/OL]. DOI: 10.48550/arXiv.1806.02473 .
|
| 13 |
NUGMANOV R I, MUKHAMETGALEEV R N, AKHMETSHIN T, et al. CGRtools: Python library for molecule, reaction, and condensed graph of reaction processing[J]. Journal of Chemical Information and Modeling, 2019, 59(6): 2516-2521.
|
| 14 |
TAVAKOLI Mohammadamin, SHMAKOV Alexander, CECCARELLI Francesco, et al. Rxn Hypergraph: A hypergraph attention model for chemical reaction representation[J/OL]. DOI: 10.48550/arXiv:2201. 01196 .
|
| 15 |
LOWE D M. Chemical reactions from US patents [P]. [2017-06-14]. .
|
| 16 |
LANDRUM G. Rdkit: Open-source chemoinformatics and machine learning. [EB/OL]. .
|
| 17 |
HU Weihua, LIU Bowen, GOMES Joseph, et al. Strategies for Pre-training Graph Neural Networks[C]//ICLR 2020, 2020.
|
| 18 |
(Reaxys Reaxys, accessed 9 May 2023) [EB/OL]. .
|
 ), DU Jian, ZHANG Lei(
), DU Jian, ZHANG Lei( )
)