化工进展 ›› 2022, Vol. 41 ›› Issue (12): 6511-6521.DOI: 10.16085/j.issn.1000-6613.2022-0418
微生物细胞工厂中代谢途径动态调控策略与网络构建
- 1.北京理工大学化学与化工学院生物化工研究所,北京 100081
2.清华大学化学工程系生物化工研究所,北京 100084
-
收稿日期:2022-03-18修回日期:2022-04-25出版日期:2022-12-20发布日期:2022-12-29 -
通讯作者:周晓宏 -
作者简介:胥健萍(1998—),女,硕士研究生,研究方向为代谢工程。E-mail:xujianping1998@163.com。 -
基金资助:国家自然科学基金(21476026)
Dynamic regulation strategies and regulation network construction of metabolic pathways in microbial cell factories
XU Jianping1( ), WANG Ying1, LI Chun1,2, ZHOU Xiaohong1(
), WANG Ying1, LI Chun1,2, ZHOU Xiaohong1( )
)
- 1.Institute of Biochemical Engineering, School of Chemistry and Chemical Engineering, Beijing Institute of Technology, Beijing 100081, China
2.Institute of Biochemical Engineering, Department of Chemical Engineering, Tsinghua University, Beijing 100084, China
-
Received:2022-03-18Revised:2022-04-25Online:2022-12-20Published:2022-12-29 -
Contact:ZHOU Xiaohong
摘要:
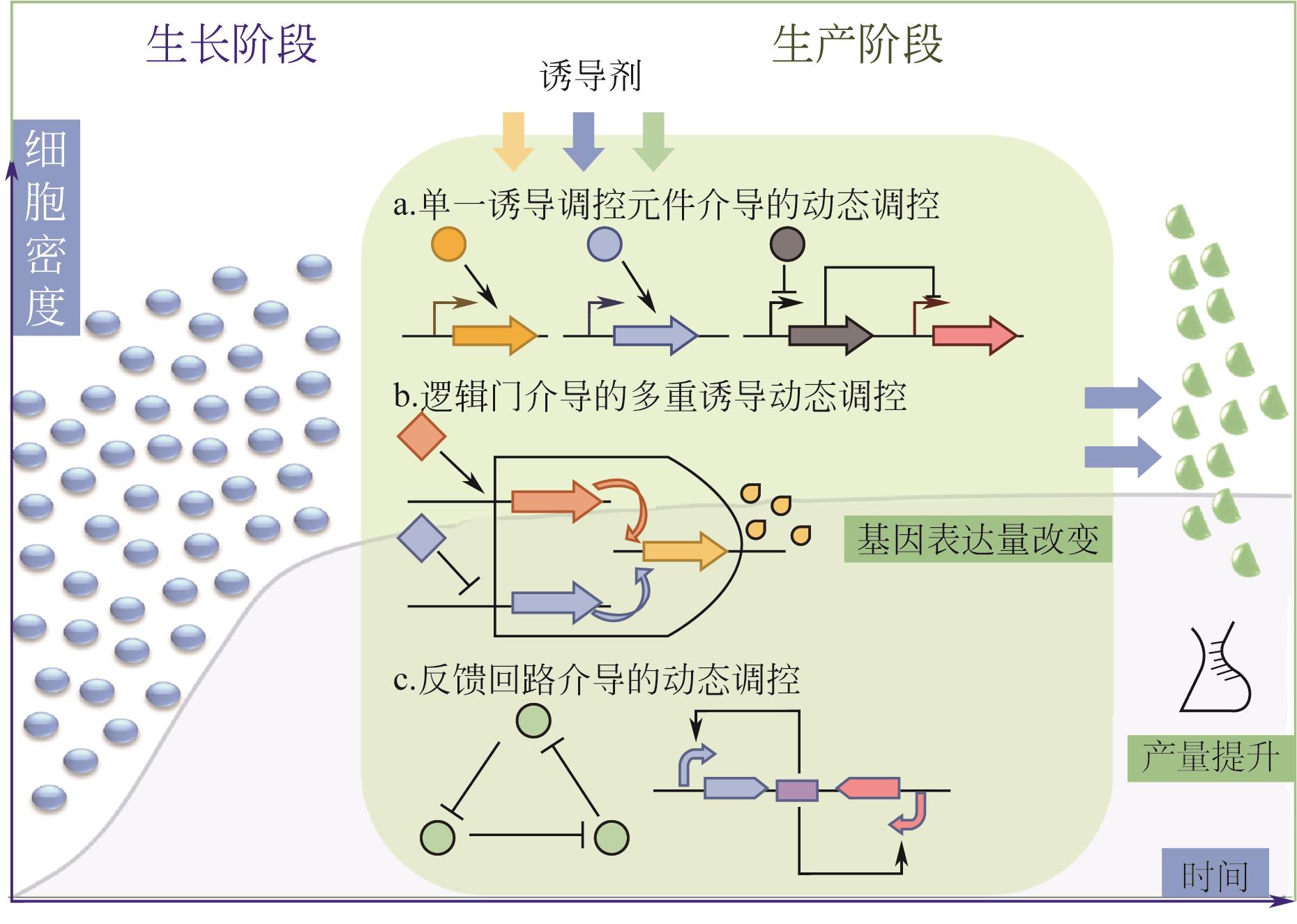
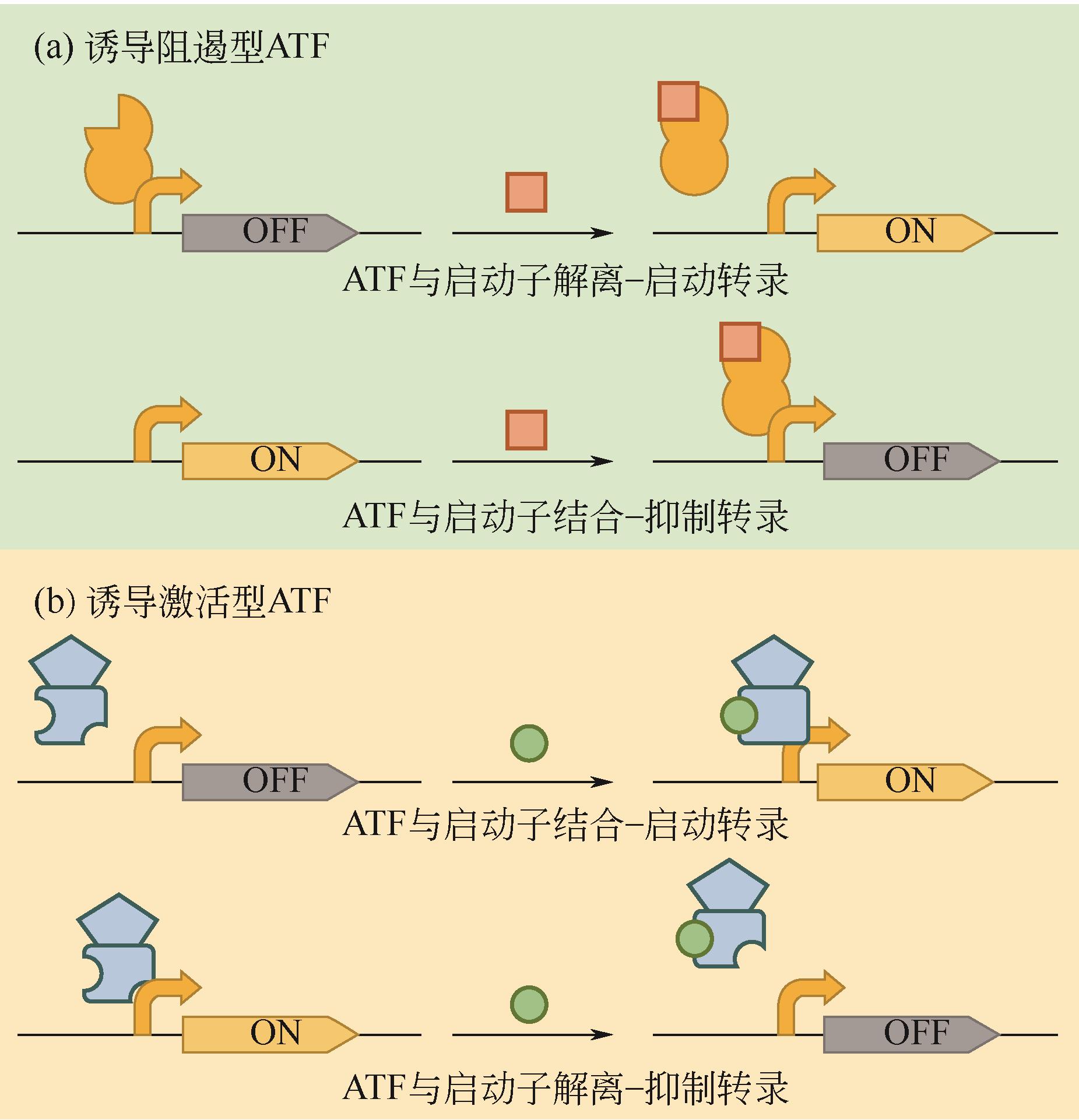
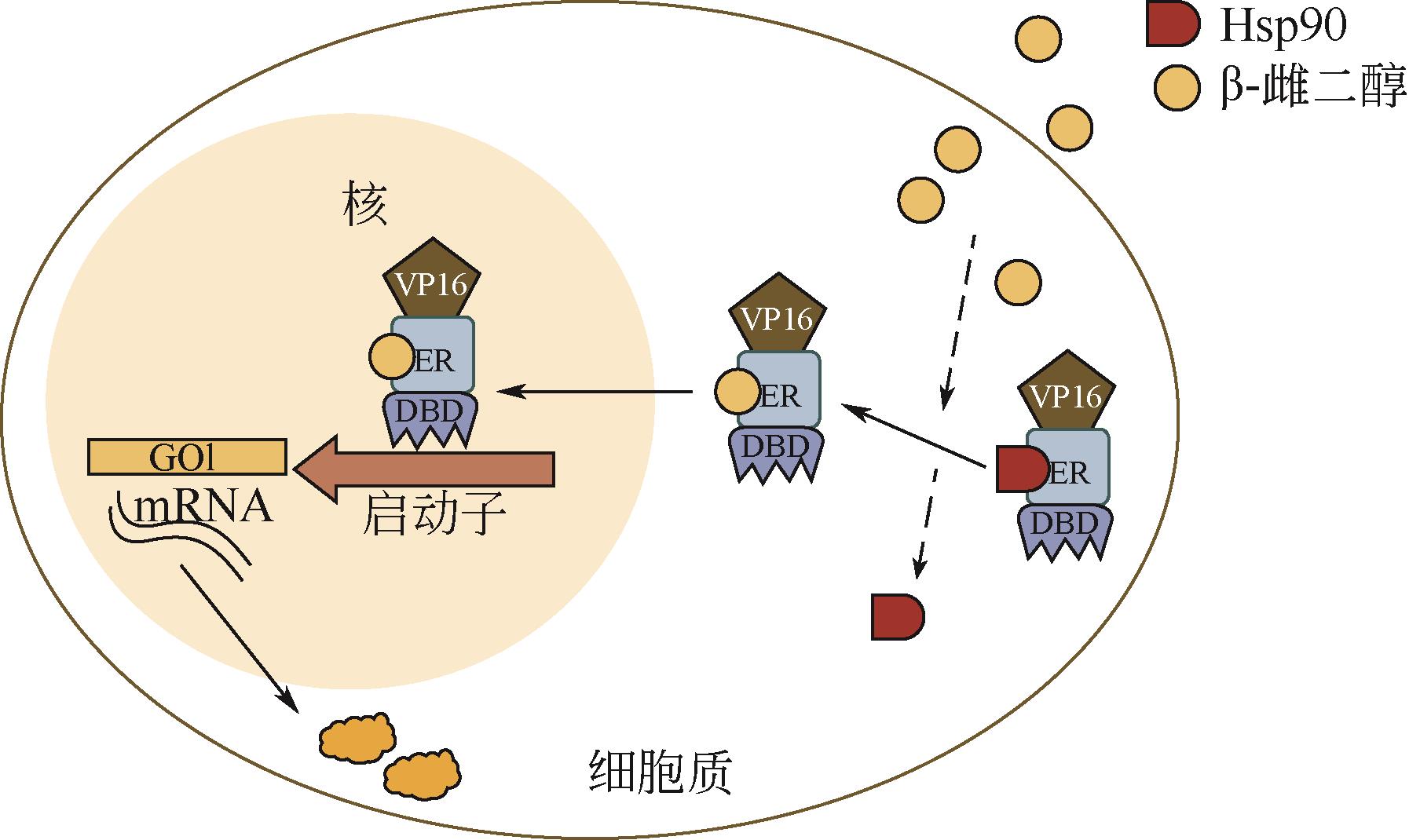
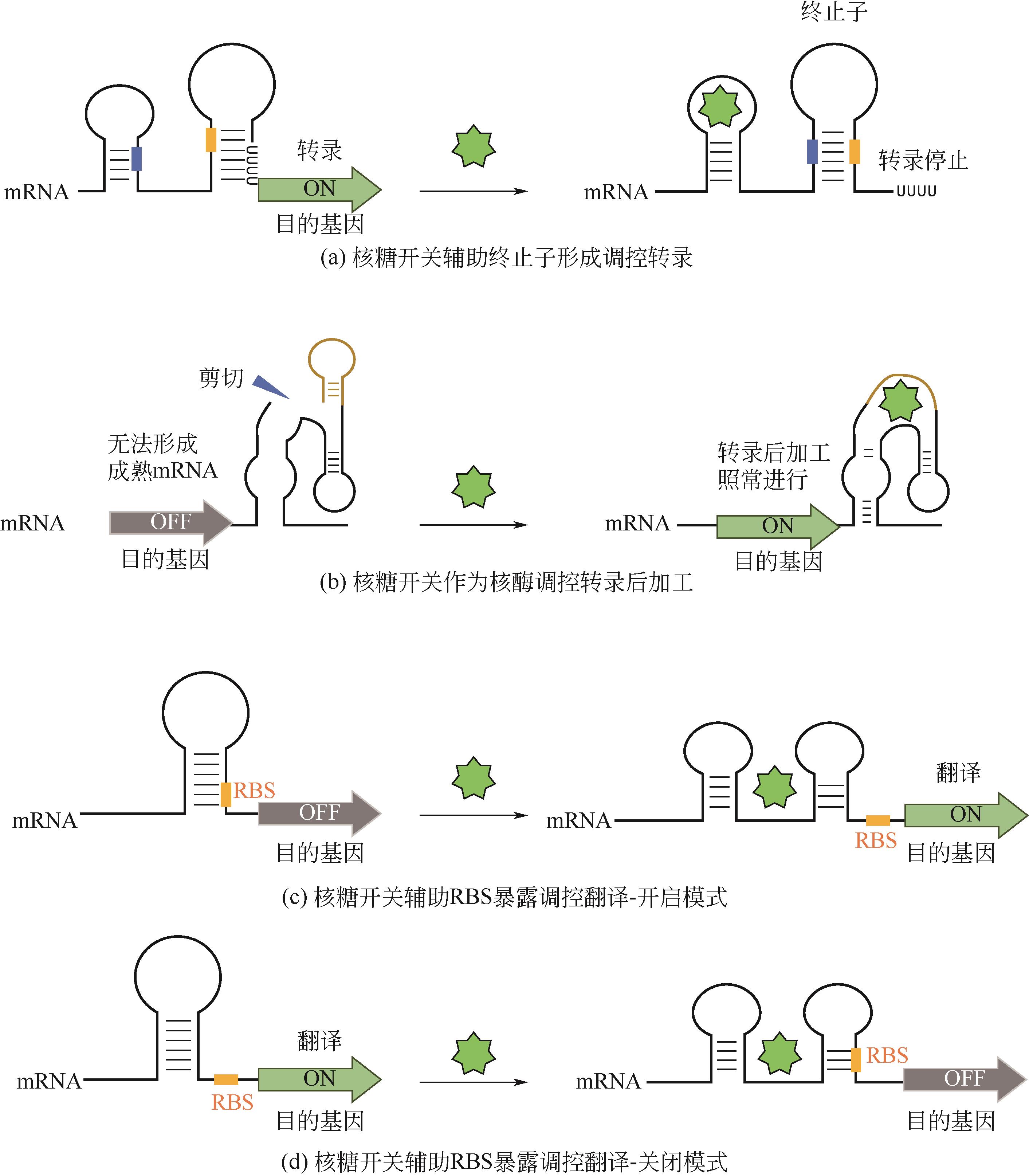
微生物细胞工厂生产目标产物时会面临营养物质消耗、代谢物积累、异源途径压力和遗传不稳定等问题,导致代谢失衡,因此需要对细胞代谢途径重新进行设计,使代谢途径根据发酵等环境条件的变化自动调节代谢通量,达到高效生产。本文首先介绍了动态调控元件的主要类型、调控机制及其应用,重点讲述了与诱导物或诱导因素作用的蛋白质转录因子调控元件和RNA核糖开关调控元件;并且从转录因子与启动子序列两个方面介绍了动态调控元件的设计与改造策略;随后总结了诱导调控元件应用于代谢途径动态调控网络构建的策略,基因表达调控已从单输入信号调控转向多输入信号逻辑门调控,通过多重诱导输入信号的逻辑门和闭环代谢物反馈回路构建更精确的动态调控网络。
中图分类号:
引用本文
胥健萍, 王颖, 李春, 周晓宏. 微生物细胞工厂中代谢途径动态调控策略与网络构建[J]. 化工进展, 2022, 41(12): 6511-6521.
XU Jianping, WANG Ying, LI Chun, ZHOU Xiaohong. Dynamic regulation strategies and regulation network construction of metabolic pathways in microbial cell factories[J]. Chemical Industry and Engineering Progress, 2022, 41(12): 6511-6521.
| 诱导剂 | 宿主 | 传感元件 | 动态 范围 | 功能 | 参考 文献 |
|---|---|---|---|---|---|
| 丙烯酸酯 | 大肠杆菌 | 转录因子AcuR | 90倍 | 阻遏 | [ |
| 红霉素 | 大肠杆菌 | 转录因子MphR | 108倍 | 阻遏 | [ |
| 柚皮素 | 大肠杆菌 | 转录因子TtgR | 70倍 | 阻遏 | [ |
| 脱水四环素 | 大肠杆菌 | 转录因子TetR | 63倍 | 阻遏 | [ |
| 香草醛 | 大肠杆菌 | 转录因子VanR | 14.2倍 | 阻遏 | [ |
| 戊二酸 | 大肠杆菌 | 转录因子CsiR | 1.5倍 | 阻遏 | [ |
| 戊二酸 | 大肠杆菌 | 转录因子CdaR | 168倍 | 激活 | [ |
| 阿拉伯糖 | 大肠杆菌 | 转录因子AraC | 210倍 | 激活 | [ |
| β-丙氨酸 | 贪铜菌吊钩虫 | 转录因子OapR | 8倍 | 激活 | [ |
| 戊二酸 | 大肠杆菌 | 转录因子GcdR | 55.5倍 | 激活 | [ |
| 雌二醇 | 酿酒酵母 | 转录因子ZEV | 50倍 | 激活 | [ |
| 蔗糖 | 酿酒酵母 | 蔗糖响应转录因子 | 2倍 | 激活 | [ |
| 铜离子 | 酿酒酵母 | 转录因子Ace1 | 6倍 | 激活 | [ |
| 赖氨酸 | 谷氨酸棒杆菌 | 赖氨酸核糖开关 | — | 阻遏 | [ |
| pH | 大肠杆菌 | pH响应核糖开关 | 31倍 | 激活 | [ |
| 四环素 | 酿酒酵母 | 四环素核糖开关 | 6倍 | 抑制 | [ |
| 硫胺素焦磷酸(TPP) | 粗糙脉孢菌 | TPP核糖开关 | 2~4倍 | 抑制 | [ |
| 茶碱 | 酿酒酵母 | 茶碱核酶开关 | 1.9倍 | 激活 | [ |
表1 应用于动态调控的基因元件
| 诱导剂 | 宿主 | 传感元件 | 动态 范围 | 功能 | 参考 文献 |
|---|---|---|---|---|---|
| 丙烯酸酯 | 大肠杆菌 | 转录因子AcuR | 90倍 | 阻遏 | [ |
| 红霉素 | 大肠杆菌 | 转录因子MphR | 108倍 | 阻遏 | [ |
| 柚皮素 | 大肠杆菌 | 转录因子TtgR | 70倍 | 阻遏 | [ |
| 脱水四环素 | 大肠杆菌 | 转录因子TetR | 63倍 | 阻遏 | [ |
| 香草醛 | 大肠杆菌 | 转录因子VanR | 14.2倍 | 阻遏 | [ |
| 戊二酸 | 大肠杆菌 | 转录因子CsiR | 1.5倍 | 阻遏 | [ |
| 戊二酸 | 大肠杆菌 | 转录因子CdaR | 168倍 | 激活 | [ |
| 阿拉伯糖 | 大肠杆菌 | 转录因子AraC | 210倍 | 激活 | [ |
| β-丙氨酸 | 贪铜菌吊钩虫 | 转录因子OapR | 8倍 | 激活 | [ |
| 戊二酸 | 大肠杆菌 | 转录因子GcdR | 55.5倍 | 激活 | [ |
| 雌二醇 | 酿酒酵母 | 转录因子ZEV | 50倍 | 激活 | [ |
| 蔗糖 | 酿酒酵母 | 蔗糖响应转录因子 | 2倍 | 激活 | [ |
| 铜离子 | 酿酒酵母 | 转录因子Ace1 | 6倍 | 激活 | [ |
| 赖氨酸 | 谷氨酸棒杆菌 | 赖氨酸核糖开关 | — | 阻遏 | [ |
| pH | 大肠杆菌 | pH响应核糖开关 | 31倍 | 激活 | [ |
| 四环素 | 酿酒酵母 | 四环素核糖开关 | 6倍 | 抑制 | [ |
| 硫胺素焦磷酸(TPP) | 粗糙脉孢菌 | TPP核糖开关 | 2~4倍 | 抑制 | [ |
| 茶碱 | 酿酒酵母 | 茶碱核酶开关 | 1.9倍 | 激活 | [ |
| 输入信号 | 宿主 | 产物 | 动态调控类型 | 产量提高 | 参考文献 |
|---|---|---|---|---|---|
| 法尼希基焦磷酸 | 大肠杆菌 | 紫穗槐二烯 | 基因开关 | 2倍 | [ |
| 丙二酰辅酶 A | 大肠杆菌 | 脂肪酸 | 基因开关 | 2.1倍 | [ |
| IPTG | 大肠杆菌 | 绿色荧光蛋白 | 基因开关 | — | [ |
| 阿拉伯糖、IPTG | 大肠杆菌 | 红色荧光蛋白 | 逻辑门(AND门) | — | [ |
| 胆汁酸、群感信号 | 大肠杆菌 | 荧光素酶 | 逻辑门(NOT/NOR) | — | [ |
| 葡萄糖、溶解氧 | 大肠杆菌 | 醋酸盐 | 逻辑门(AND/NAND) | 4倍 | [ |
| IPTG、群感信号、阿拉伯糖 | 大肠杆菌 | 黄色荧光蛋白 | 逻辑门(NOR/NOT) | — | [ |
| IPTG | 大肠杆菌 | 脂肪酸 | 反馈回路 | 12倍 | [ |
| n-乙酰葡糖胺 | 枯草芽孢杆菌 | n-乙酰葡糖胺 | 反馈回路 | 2.19倍 | [ |
| 法尼基焦磷酸 | 大肠杆菌 | 紫穗槐二烯 | 反馈回路 | 2倍 | [ |
| 丙二酰辅酶A | 大肠杆菌 | 脂肪酸 | 反馈回路 | 2倍 | [ |
表2 代谢途径的动态调控网络构建
| 输入信号 | 宿主 | 产物 | 动态调控类型 | 产量提高 | 参考文献 |
|---|---|---|---|---|---|
| 法尼希基焦磷酸 | 大肠杆菌 | 紫穗槐二烯 | 基因开关 | 2倍 | [ |
| 丙二酰辅酶 A | 大肠杆菌 | 脂肪酸 | 基因开关 | 2.1倍 | [ |
| IPTG | 大肠杆菌 | 绿色荧光蛋白 | 基因开关 | — | [ |
| 阿拉伯糖、IPTG | 大肠杆菌 | 红色荧光蛋白 | 逻辑门(AND门) | — | [ |
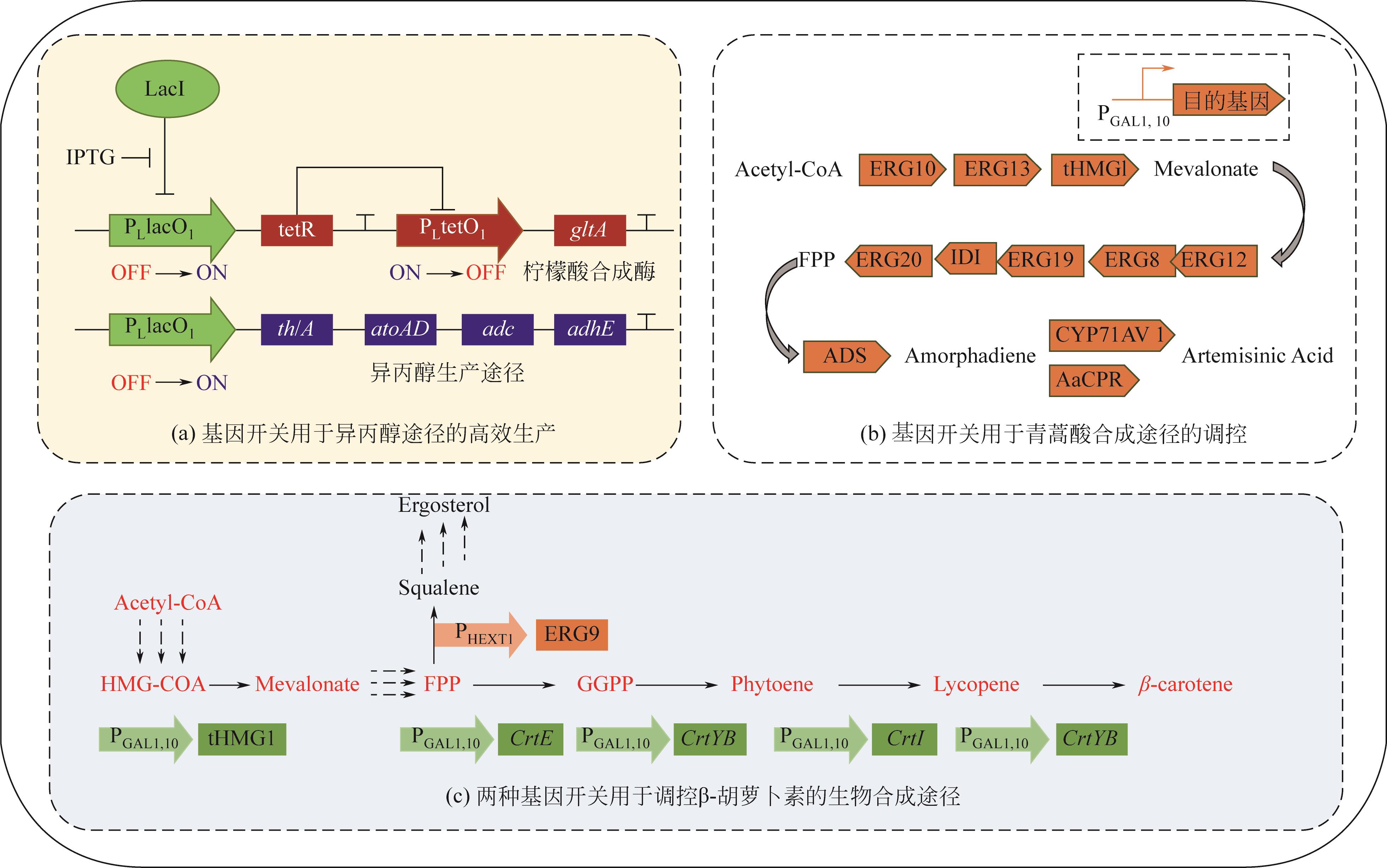
| 胆汁酸、群感信号 | 大肠杆菌 | 荧光素酶 | 逻辑门(NOT/NOR) | — | [ |
| 葡萄糖、溶解氧 | 大肠杆菌 | 醋酸盐 | 逻辑门(AND/NAND) | 4倍 | [ |
| IPTG、群感信号、阿拉伯糖 | 大肠杆菌 | 黄色荧光蛋白 | 逻辑门(NOR/NOT) | — | [ |
| IPTG | 大肠杆菌 | 脂肪酸 | 反馈回路 | 12倍 | [ |
| n-乙酰葡糖胺 | 枯草芽孢杆菌 | n-乙酰葡糖胺 | 反馈回路 | 2.19倍 | [ |
| 法尼基焦磷酸 | 大肠杆菌 | 紫穗槐二烯 | 反馈回路 | 2倍 | [ |
| 丙二酰辅酶A | 大肠杆菌 | 脂肪酸 | 反馈回路 | 2倍 | [ |
| 1 | CHOI Yong Jun, LEE Sang Yup. Microbial production of short-chain alkanes[J]. Nature, 2013, 502(7472): 571-574. |
| 2 | SHEPPARD Micah J, KUNJAPUR Aditya M, WENCK Spencer J, et al. Retro-biosynthetic screening of a modular pathway design achieves selective route for microbial synthesis of 4-methyl-pentanol[J]. Nature Communications, 2014, 5: 5031. |
| 3 | FEI Qiang, GUARNIERI Michael T, TAO Ling, et al. Bioconversion of natural gas to liquid fuel: opportunities and challenges[J]. Biotechnology Advances, 2014, 32(3): 596-614. |
| 4 | NIELSEN Jens. Cell factory engineering for improved production of natural products[J]. Natural Product Reports, 2019, 36(9): 1233-1236. |
| 5 | CHEN Ruibing, YANG Shan, ZHANG Lei, et al. Advanced strategies for production of natural products in yeast[J]. iScience, 2020, 23(3): 100879. |
| 6 | ZHOU Yongjin J, KERKHOVEN Eduard J, NIELSEN Jens. Barriers and opportunities in bio-based production of hydrocarbons[J]. Nature Energy, 2018, 3(11): 925-935. |
| 7 | BENTLEY Gayle J, JIANG Wen, GUAMÁN Linda P, et al. Engineering Escherichia coli to produce branched-chain fatty acids in high percentages[J]. Metabolic Engineering, 2016, 38: 148-158. |
| 8 | CHARUSANTI Pep, CONRAD Tom M, KNIGHT Eric M, et al. Genetic basis of growth adaptation of Escherichia coli after deletion of pgi, a major metabolic gene[J]. PLoS Genetics, 2010, 6(11): e1001186. |
| 9 | KIZER Lance, PITERA Douglas J, PFLEGER Brian F, et al. Application of functional genomics to pathway optimization for increased isoprenoid production[J]. Applied and Environmental Microbiology, 2008, 74(10): 3229-3241. |
| 10 | LIU Di, MANNAN Ahmad A, HAN Yichao, et al. Dynamic metabolic control: towards precision engineering of metabolism[J]. Journal of Industrial Microbiology & Biotechnology, 2018, 45(7): 535-543. |
| 11 | LIAN Jiazhang, MISHRA Shekhar, ZHAO Huimin. Recent advances in metabolic engineering of Saccharomyces cerevisiae: new tools and their applications[J]. Metabolic Engineering, 2018, 50: 85-108. |
| 12 | KOCHANOWSKI Karl, GEROSA Luca, BRUNNER Simon F, et al. Few regulatory metabolites coordinate expression of central metabolic genes in Escherichia coli [J]. Molecular Systems Biology, 2017, 13(1): 903. |
| 13 | CHUBUKOV Victor, GEROSA Luca, KOCHANOWSKI Karl, et al. Coordination of microbial metabolism[J]. Nature Reviews Microbiology, 2014, 12(5): 327-340. |
| 14 | CHIN Chen-Shan, CHUBUKOV Victor, JOLLY Emmitt R, et al. Dynamics and design principles of a basic regulatory architecture controlling metabolic pathways[J]. PLoS Biology, 2008, 6(6): e146. |
| 15 | JOZEFCZUK Szymon, KLIE Sebastian, CATCHPOLE Gareth, et al. Metabolomic and transcriptomic stress response of Escherichia coli [J]. Molecular Systems Biology, 2010, 6: 364. |
| 16 | ANESIADIS Nikolaos, CLUETT William R, MAHADEVAN Radhakrishnan. Dynamic metabolic engineering for increasing bioprocess productivity[J]. Metabolic Engineering, 2008, 10(5): 255-266. |
| 17 | CRESS Brady F, TRANTAS Emmanouil A, VERVERIDIS Filippos, et al. Sensitive cells: enabling tools for static and dynamic control of microbial metabolic pathways[J]. Current Opinion in Biotechnology, 2015, 36: 205-214. |
| 18 | DAHL Robert H, ZHANG Fuzhong, Jorge ALONSO-GUTIERREZ, et al. Engineering dynamic pathway regulation using stress-response promoters[J]. Nature Biotechnology, 2013, 31(11): 1039-1046. |
| 19 | ROGERS Jameson K, GUZMAN Christopher D, TAYLOR Noah D, et al. Synthetic biosensors for precise gene control and real-time monitoring of metabolites[J]. Nucleic Acids Research, 2015, 43(15): 7648-7660. |
| 20 | KUNJAPUR Aditya M, PRATHER Kristala L J. Development of a vanillate biosensor for the vanillin biosynthesis pathway in E. coli [J]. ACS Synthetic Biology, 2019, 8(9): 1958-1967. |
| 21 | THOMPSON Mitchell G, COSTELLO Zak, HUMMEL Niklas F C, et al. Robust characterization of two distinct glutarate sensing transcription factors of Pseudomonas putida L-lysine metabolism[J]. ACS Synthetic Biology, 2019, 8(10): 2385-2396. |
| 22 | ROGERS Jameson K, CHURCH George M. Genetically encoded sensors enable real-time observation of metabolite production[J]. Proceedings of the National Academy of Sciences of the United States of America, 2016, 113(9): 2388-2393. |
| 23 | HANKO Erik K R, PAIVA Ana C, JONCZYK Magdalena, et al. A genome-wide approach for identification and characterisation of metabolite-inducible systems[J]. Nature Communications, 2020, 11(1): 1213. |
| 24 | Scott MCISAAC R, OAKES Benjamin L, WANG Xin, et al. Synthetic gene expression perturbation systems with rapid, tunable, single-gene specificity in yeast[J]. Nucleic Acids Research, 2013, 41(4): e57. |
| 25 | WILLIAMS Thomas C, ESPINOSA Monica I, NIELSEN Lars K, et al. Dynamic regulation of gene expression using sucrose responsive promoters and RNA interference in Saccharomyces cerevisiae [J]. Microbial Cell Factories, 2015, 14: 43. |
| 26 | KIM Sin Il, Byung Suk HA, KIM Min Seek, et al. Evaluation of copper-inducible fungal laccase promoter in foreign gene expression in Pichia pastoris [J]. Biotechnology and Bioprocess Engineering, 2016, 21(1): 53-59. |
| 27 | ZHOU Libang, ZENG A. Exploring lysine riboswitch for metabolic flux control and improvement of L-lysine synthesis in Corynebacterium glutamicum [J]. ACS Synthetic Biology, 2015, 4(6): 729-734. |
| 28 | PHAM Hoang Long, WONG Adison, CHUA Niying, et al. Engineering a riboswitch-based genetic platform for the self-directed evolution of acid-tolerant phenotypes[J]. Nature Communications, 2017, 8: 411. |
| 29 | SUESS Beatrix, HANSON Shane, BERENS Christian, et al. Conditional gene expression by controlling translation with tetracycline-binding aptamers[J]. Nucleic Acids Research, 2003, 31(7): 1853-1858. |
| 30 | CHEAH Ming T, WACHTER Andreas, SUDARSAN Narasimhan, et al. Control of alternative RNA splicing and gene expression by eukaryotic riboswitches[J]. Nature, 2007, 447(7143): 497-500. |
| 31 | BABISKIN Andrew H, SMOLKE Christina D. Engineering ligand-responsive RNA controllers in yeast through the assembly of RNase Ⅲ tuning modules[J]. Nucleic Acids Research, 2011, 39(12): 5299-5311. |
| 32 | ZHANG Liyuan, GUO Wei, LU Yuan. Advances in cell-free biosensors: principle, mechanism, and applications[J]. Biotechnology Journal, 2020, 15(9): e2000187. |
| 33 | SHEN Xiaolin, WANG Jia, LI Chenyi, et al. Dynamic gene expression engineering as a tool in pathway engineering[J]. Current Opinion in Biotechnology, 2019, 59: 122-129. |
| 34 | DING Nana, ZHOU Shenghu, DENG Yu. Transcription-factor-based biosensor engineering for applications in synthetic biology[J]. ACS Synthetic Biology, 2021, 10(5): 911-922. |
| 35 | WAN Xia, MARSAFARI Monireh, XU Peng. Engineering metabolite-responsive transcriptional factors to sense small molecules in eukaryotes: current state and perspectives[J]. Microbial Cell Factories, 2019, 18(1): 61. |
| 36 | MANNAN Ahmad A, LIU Di, ZHANG Fuzhong, et al. Fundamental design principles for transcription-factor-based metabolite biosensors[J]. ACS Synthetic Biology, 2017, 6(10): 1851-1859. |
| 37 | XU Peng. Production of chemicals using dynamic control of metabolic fluxes[J]. Current Opinion in Biotechnology, 2018, 53: 12-19. |
| 38 | SAEKI Kazuya, TOMINAGA Masahiro, KAWAI-NOMA S, et al. Rapid diversification of BetI-based transcriptional switches for the control of biosynthetic pathways and genetic circuits[J]. ACS Synthetic Biology, 2016, 5(11): 1201-1210. |
| 39 | XU Peng, LI Lingyun, ZHANG Fuming, et al. Improving fatty acids production by engineering dynamic pathway regulation and metabolic control[J]. Proceedings of the National Academy of Sciences of the United States of America, 2014, 111(31): 11299-11304. |
| 40 | HANKO Erik K R, MINTON Nigel P, MALYS Naglis. A transcription factor-based biosensor for detection of itaconic acid[J]. ACS Synthetic Biology, 2018, 7(5): 1436-1446. |
| 41 | MAHR Regina, FRUNZKE Julia. Transcription factor-based biosensors in biotechnology: current state and future prospects[J]. Applied Microbiology and Biotechnology, 2016, 100(1): 79-90. |
| 42 | WANG Baojun, BARAHONA Mauricio, BUCK Martin. Amplification of small molecule-inducible gene expression via tuning of intracellular receptor densities[J]. Nucleic Acids Research, 2015, 43(3): 1955-1964. |
| 43 | CHOU Howard H, KEASLING Jay D. Programming adaptive control to evolve increased metabolite production[J]. Nature Communications, 2013, 4: 2595. |
| 44 | XIE Wenping, YE Lidan, Xiaomei LYU, et al. Sequential control of biosynthetic pathways for balanced utilization of metabolic intermediates in Saccharomyces cerevisiae [J]. Metabolic Engineering, 2015, 28: 8-18. |
| 45 | PADDON C J, WESTFALL P J, PITERA D J, et al. High-level semi-synthetic production of the potent antimalarial artemisinin[J]. Nature, 2013, 496(7446): 528-532. |
| 46 | WEI Wenping, SHANG Yanzhe, ZHANG Ping, et al. Engineering prokaryotic transcriptional activator XylR as a xylose-inducible biosensor for transcription activation in yeast[J]. ACS Synthetic Biology, 2020, 9(5): 1022-1029. |
| 47 | RODRIGUES A, BECKER Judith, DE SOUZA LIMA A O, et al. Systems metabolic engineering of Escherichia coli for gram scale production of the antitumor drug deoxyviolacein from glycerol[J]. Biotechnology and Bioengineering, 2014, 111(11): 2280-2289. |
| 48 | ZHANG Yanfei, CORTEZ Jeremy D, HAMMER Sarah K, et al. Biosensor for branched-chain amino acid metabolism in yeast and applications in isobutanol and isopentanol production[J]. Nature Communications, 2022, 13(1): 270. |
| 49 | DIETRICH J, SHIS D, ALIKHANI A, et al. Transcription factor-based screens and synthetic selections for microbial small-molecule biosynthesis[J]. ACS synthetic biology, 2013, 2: 47-58. |
| 50 | SIEDLER Solvej, STAHLHUT Steen G, MALLA Sailesh, et al. Novel biosensors based on flavonoid-responsive transcriptional regulators introduced into Escherichia coli [J]. Metabolic Engineering, 2014, 21: 2-8. |
| 51 | RUTHERFORD Julian C, BIRD Amanda J. Metal-responsive transcription factors that regulate iron, zinc, and copper homeostasis in eukaryotic cells[J]. Eukaryotic Cell, 2004, 3(1): 1-13. |
| 52 | XIE Wenping, LIU Min, Xiaomei LYU, et al. Construction of a controllable β‐carotene biosynthetic pathway by decentralized assembly strategy in Saccharomyces cerevisiae [J]. Biotechnology and Bioengineering, 2014, 111(1): 125-133. |
| 53 | PENG Bingyin, PLAN Manuel R, CARPENTER Alexander, et al. Coupling gene regulatory patterns to bioprocess conditions to optimize synthetic metabolic modules for improved sesquiterpene production in yeast[J]. Biotechnology for Biofuels, 2017, 10(1): 1-16. |
| 54 | LIU Junfeng, WANG Tao, JIANG Yixun, et al. Harnessing β-estradiol inducible expression system to overproduce nervonic acid in Saccharomyces cerevisiae [J]. Process Biochemistry, 2020, 92: 37-42. |
| 55 | KIRBY J, ROMANINI Dante W, PARADISE Eric M, et al. Engineering triterpene production in Saccharomyces cerevisiae–beta‐amyrin synthase from Artemisia annua [J]. The FEBS Journal, 2008, 275(8): 1852-1859. |
| 56 | QIU Chenxi, CHEN Xiaoxu, REXIDA Reheman, et al. Engineering transcription factor-based biosensors for repressive regulation through transcriptional deactivation design in Saccharomyces cerevisiae [J]. Microbial Cell Factories, 2020, 19(1): 146. |
| 57 | SERGANOV Alexander, NUDLER Evgeny. A decade of riboswitches[J]. Cell, 2013, 152(1/2): 17-24. |
| 58 | MISHLER Dennis M, GALLIVAN Justin P. A family of synthetic riboswitches adopts a kinetic trapping mechanism[J]. Nucleic Acids Research, 2014, 42(10): 6753-6761. |
| 59 | REYNOSO Colleen M K, MILLER Mark A, BINA James E, et al. Riboswitches for intracellular study of genes involved in Francisella pathogenesis[J]. mBio, 2012, 3(6): e00253-e00212. |
| 60 | LYNCH Sean A, GALLIVAN Justin P. A flow cytometry-based screen for synthetic riboswitches[J]. Nucleic Acids Research, 2008, 37(1): 184-192. |
| 61 | LIANG Joe C, CHANG Andrew L, KENNEDY Andrew B, et al. A high-throughput, quantitative cell-based screen for efficient tailoring of RNA device activity[J]. Nucleic Acids Research, 2012, 40(20): e154. |
| 62 | Maung Nyan WIN, SMOLKE Christina D. Higher-order cellular information processing with synthetic RNA devices[J]. Science, 2008, 322(5900): 456-460. |
| 63 | SARAGLIADIS Athanasios, HARTIG Jörg S. Ribozyme-based transfer RNA switches for post-transcriptional control of amino acid identity in protein synthesis[J]. Journal of the American Chemical Society, 2013, 135(22): 8222-8226. |
| 64 | BERSCHNEIDER Barbara, WIELAND Markus, RUBINI Marina, et al. Small-molecule-dependent regulation of transfer RNA in bacteria[J]. Angewandte Chemie International Edition, 2009, 48(41): 7564-7567. |
| 65 | OGAWA Atsushi, MAEDA Mizuo. A novel label-free biosensor using an aptazyme-suppressor-tRNA conjugate and an amber mutated reporter gene[J]. Chembiochem, 2008, 9(14): 2204-2208. |
| 66 | MAHR Regina, Cornelia GÄTGENS, Jochem GÄTGENS, et al. Biosensor-driven adaptive laboratory evolution of L-valine production in Corynebacterium glutamicum [J]. Metabolic Engineering, 2015, 32: 184-194. |
| 67 | ZHOU Yikang, YUAN Yaomeng, WU Yinan, et al. Encoding genetic circuits with DNA barcodes paves the way for machine learning-assisted metabolite biosensor response curve profiling in yeast[J]. ACS Synthetic Biology, 2022, 11(2): 977-989. |
| 68 | XU Ning, WEI Liang, LIU Jun. Recent advances in the applications of promoter engineering for the optimization of metabolite biosynthesis[J]. World Journal of Microbiology & Biotechnology, 2019, 35(2): 33. |
| 69 | Keith E J TYO, NEVOIGT Elke, STEPHANOPOULOS Gregory. Directed evolution of promoters and tandem gene arrays for customizing RNA synthesis rates and regulation[M]//Methods in Enzymology. Amsterdam: Elsevier, 2011: 135-155. |
| 70 | WEI Liang, XU Ning, WANG Yiran, et al. Promoter library-based module combination (PLMC) technology for optimization of threonine biosynthesis in Corynebacterium glutamicum [J]. Applied Microbiology and Biotechnology, 2018, 102(9): 4117-4130. |
| 71 | BLAZECK John, GARG Rishi, REED Ben, et al. Controlling promoter strength and regulation in Saccharomyces cerevisiae using synthetic hybrid promoters[J]. Biotechnology and Bioengineering, 2012, 109(11): 2884-2895. |
| 72 | BLAZECK John, LIU Leqian, REDDEN Heidi, et al. Tuning gene expression in Yarrowia lipolytica by a hybrid promoter approach[J]. Applied and Environmental Microbiology, 2011, 77(22): 7905-7914. |
| 73 | REDDEN Heidi, ALPER Hal S. The development and characterization of synthetic minimal yeast promoters[J]. Nature Communications, 2015, 6: 7810. |
| 74 | MURPHY K F, BALÁZSI G, COLLINS J J. Combinatorial promoter design for engineering noisy gene expression[J]. Proceedings of the National Academy of Sciences of the United States of America, 2007, 104(31): 12726-12731. |
| 75 | XU Peng, WANG Wenya, LI Lingyun, et al. Design and kinetic analysis of a hybrid promoter–regulator system for malonyl-CoA sensing in Escherichia coli [J]. ACS Chemical Biology, 2014, 9(2): 451-458. |
| 76 | GARDNER Timothy S, CANTOR Charles R, COLLINS James J. Construction of a genetic toggle switch in Escherichia coli [J]. Nature, 2000, 403(6767): 339-342. |
| 77 | MOON Tae Seok, LOU Chunbo, TAMSIR Alvin, et al. Genetic programs constructed from layered logic gates in single cells[J]. Nature, 2012, 491(7423): 249-253. |
| 78 | TAKETANI Mao, ZHANG Jianbo, ZHANG Shuyi, et al. Genetic circuit design automation for the gut resident species Bacteroides thetaiotaomicron [J]. Nature Biotechnology, 2020, 38(8): 962-969. |
| 79 | MOSER Felix, BORUJENI Amin Espah, GHODASARA Amar N, et al. Dynamic control of endogenous metabolism with combinatorial logic circuits[J]. Molecular Systems Biology, 2018, 14(11): e8605. |
| 80 | GOROCHOWSKI Thomas E, BORUJENI Amin Espah, PARK Yongjin, et al. Genetic circuit characterization and debugging using RNA-seq[J]. Molecular Systems Biology, 2017, 13(11): 952. |
| 81 | LIU Di, ZHANG Fuzhong. Metabolic feedback circuits provide rapid control of metabolite dynamics[J]. ACS Synthetic Biology, 2018, 7(2): 347-356. |
| 82 | WU Yaokang, CHEN Taichi, LIU Yanfeng, et al. Design of a programmable biosensor-CRISPRi genetic circuits for dynamic and autonomous dual-control of metabolic flux in Bacillus subtilis [J]. Nucleic Acids Research, 2020, 48(2): 996-1009. |
| 83 | BROCKMAN Irene M, PRATHER Kristala L J. Dynamic knockdown of E. coli central metabolism for redirecting fluxes of primary metabolites[J]. Metabolic Engineering, 2015, 28: 104-113. |
| 84 | SOMA Yuki, YAMAJI Taiki, MATSUDA Fumio, et al. Synthetic metabolic bypass for a metabolic toggle switch enhances acetyl-CoA supply for isopropanol production by Escherichia coli [J]. Journal of Bioscience and Bioengineering, 2017, 123(5): 625-633. |
| 85 | WESTFALL P J, PITERA D J, LENIHAN J R, et al. Production of amorphadiene in yeast, and its conversion to dihydroartemisinic acid, precursor to the antimalarial agent artemisinin[J]. Proceedings of the National Academy of Sciences of the United States of America, 2012, 109(3): E111-E118. |
| 86 | ANDERSON J C, VOIGT C A, ARKIN A P. Environmental signal integration by a modular AND gate[J]. Molecular Systems Biology, 2007, 3: 133. |
| 87 | Hyun Gyu LIM, Myung Hyun NOH, JEONG Jun Hong, et al. Optimum rebalancing of the 3-hydroxypropionic acid production pathway from glycerol in Escherichia coli [J]. ACS Synthetic Biology, 2016, 5(11): 1247-1255. |
| 88 | ZHOU Kang, QIAO Kangjian, EDGAR Steven, et al. Distributing a metabolic pathway among a microbial consortium enhances production of natural products[J]. Nature Biotechnology, 2015, 33(4): 377-383. |
| 89 | LARA Alvaro R, JAÉN Karim E, SIGALA Juan-Carlos, et al. Characterization of endogenous and reduced promoters for oxygen-limited processes using Escherichia coli [J]. ACS Synthetic Biology, 2017, 6(2): 344-356. |
| 90 | MAHR Regina, BOESELAGER Raphael Freiherr, WIECHERT Johanna, et al. Screening of an Escherichia coli promoter library for a phenylalanine biosensor[J]. Applied Microbiology and Biotechnology, 2016, 100(15): 6739-6753. |
| 91 | SNOEK Tim, CHABERSKI Evan K, AMBRI Francesca, et al. Evolution-guided engineering of small-molecule biosensors[J]. Nucleic Acids Research, 2020, 48(1): e3. |
| 92 | SKJOEDT Mette L, SNOEK Tim, KILDEGAARD Kanchana R, et al. Engineering prokaryotic transcriptional activators as metabolite biosensors in yeast[J]. Nature Chemical Biology, 2016, 12(11): 951-958. |
| 93 | YEOH Jing Wui, Kai Boon Ivan NG, Ai Ying TEH, et al. An automated biomodel selection system (BMSS) for gene circuit designs[J]. ACS Synthetic Biology, 2019, 8(7): 1484-1497. |
| 94 | LIAO Chen, BLANCHARD Andrew E, LU Ting. An integrative circuit-host modelling framework for predicting synthetic gene network behaviours[J]. Nature Microbiology, 2017, 2(12): 1658-1666. |
| [1] | 刘情, 王小婉, 诸葛斌, 陆信曜, 宗红, 方慧英, 宋健. 基因敲除弱化产1,3-丙二醇Klebsiella pneumoniae荚膜多糖的合成[J]. 化工进展, 2017, 36(09): 3447-3452. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||