化工进展 ›› 2019, Vol. 38 ›› Issue (05): 2402-2412.DOI: 10.16085/j.issn.1000-6613.2018-1637
高通量筛选技术在菌种进化中的研究进展
杨祖明1,2( ),王颖1,2,姚明东1,2(
),王颖1,2,姚明东1,2( ),肖文海1,2
),肖文海1,2
- 1. 天津大学系统生物工程教育部重点实验室,天津 300072
2. 天津化学化工协同创新中心合成生物学平台,天津 300072
-
收稿日期:2018-08-09修回日期:2018-10-16出版日期:2019-05-05发布日期:2019-05-05 -
通讯作者:姚明东 -
作者简介:<named-content content-type="corresp-name">杨祖明</named-content>(1993—),男,硕士研究生,研究方向为合成生物学。E-mail:<email>zumingyang@tju.edu.cn</email>。|姚明东,副研究员,研究方向为细胞工程与酶工程。E-mail:<email>mingdong.yao@tju.edu.cn</email>。 -
基金资助:国家自然科学基金青年基金项目(31600052);天津市创新平台与人才项目(16PTSYJC00050和16PTGCCX00140)
High-throughput screening technology in strain evolution
Zuming YANG1,2( ),Ying WANG1,2,Mingdong YAO1,2(
),Ying WANG1,2,Mingdong YAO1,2( ),Wenhai XIAO1,2
),Wenhai XIAO1,2
- 1. Key Laboratory of Systems Bioengineering (Ministry of Education), Tianjin University, Tianjin 300072, China
2. SynBio Research Platform, Collaborative Innovation Center of Chemical Science and Engineering, Tianjin 300072, China
-
Received:2018-08-09Revised:2018-10-16Online:2019-05-05Published:2019-05-05 -
Contact:Mingdong YAO
摘要:
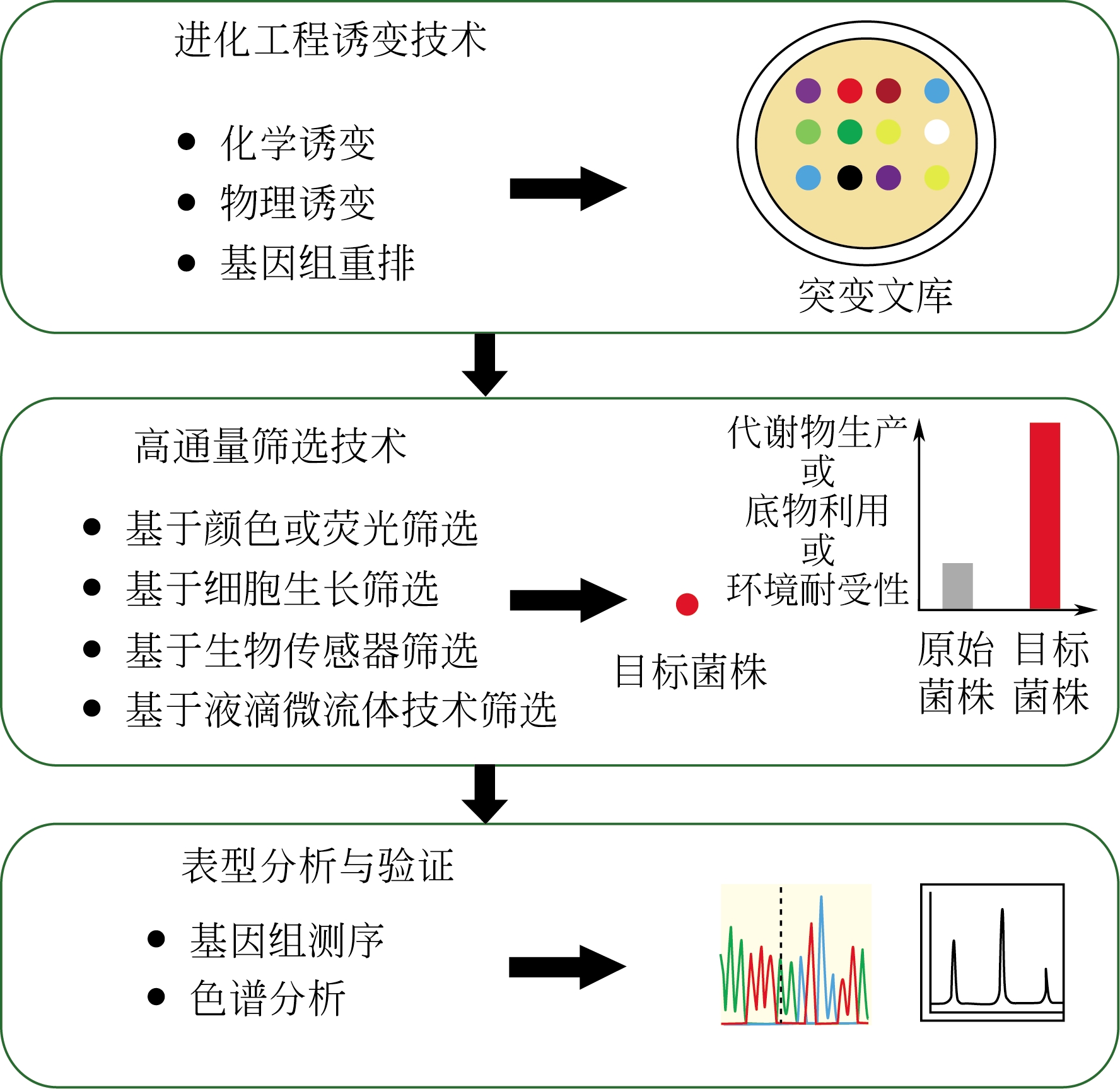
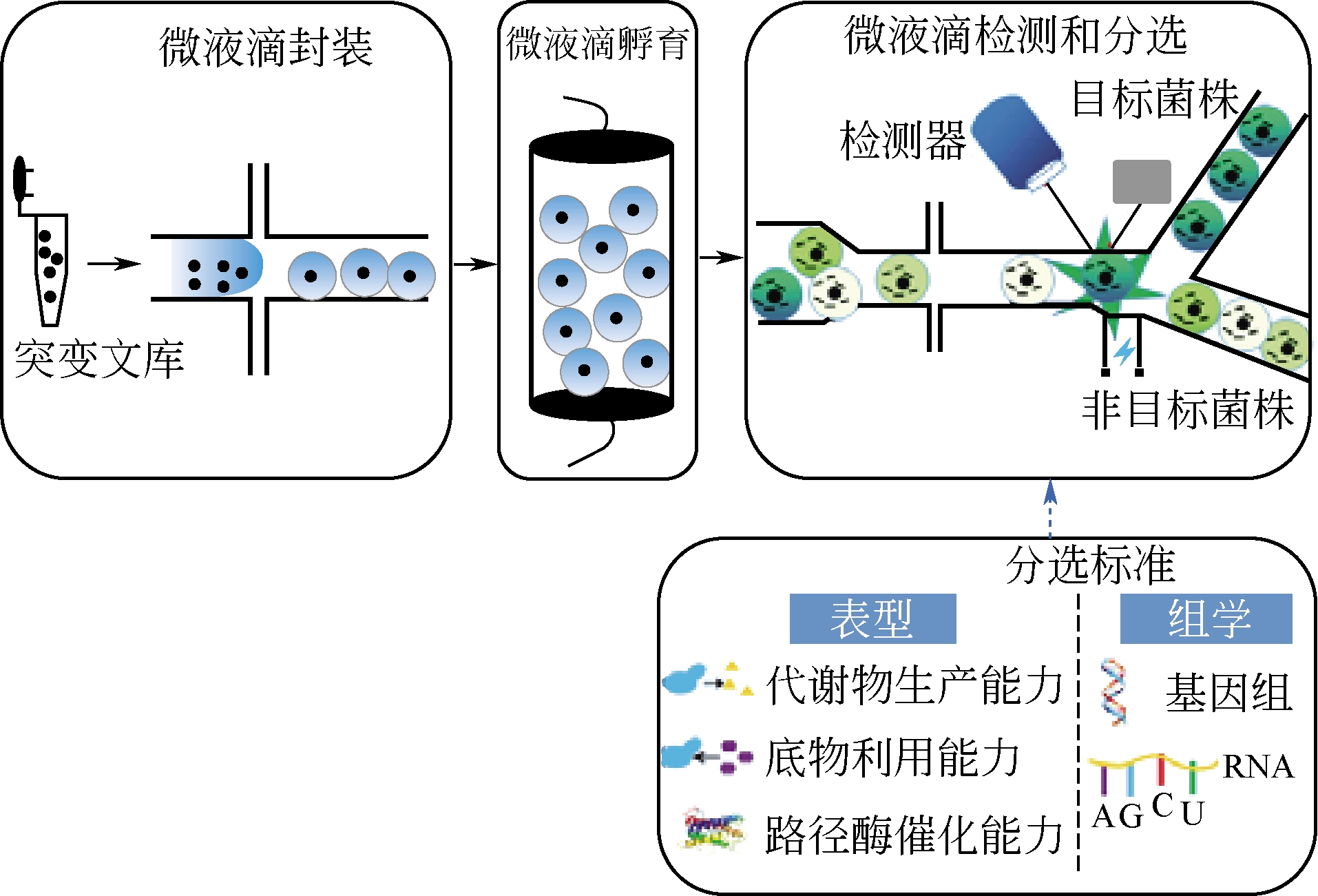
菌种进化工程是绿色生物制造的重要策略,利用高效的高通量筛选方法和技术可以快速地获得理想的实用菌株。针对菌种进化工程中的高通量筛选方法,本文重点综述了基于颜色或荧光、基于细胞生长、基于生物传感器以及基于液滴微流体平台等4个方面的高通量筛选技术的重要进展,同时也介绍了各种高通量筛选技术的应用范围和特点,为研究人员从不同进化文库中获得生理特性或者代谢能力显著提高的目标菌株提供了理论指导,极大地提高进化文库的筛选效率,降低了菌株筛选的时间和成本。最后展望了人工智能、合成生物学以及生物信息学的发展对高通量筛选技术的重要影响,以期提高高通量筛选技术的精度、效率和应用范围,进而加速菌种进化过程和工业化进程。
中图分类号:
引用本文
杨祖明, 王颖, 姚明东, 肖文海. 高通量筛选技术在菌种进化中的研究进展[J]. 化工进展, 2019, 38(05): 2402-2412.
Zuming YANG, Ying WANG, Mingdong YAO, Wenhai XIAO. High-throughput screening technology in strain evolution[J]. Chemical Industry and Engineering Progress, 2019, 38(05): 2402-2412.
| 菌株 | 进化手段 | 进化目的 | 突变文库大小 | 筛选方法 | 参考文献 |
|---|---|---|---|---|---|
| 莱茵衣藻 | 基因重组 | 提高细胞脂质积累 | 6×104 | 基于细胞颜色筛选 | [ |
| 酵母 | UV诱变 | 提高菌株α-淀粉酶的分泌能力 | 105 | 液滴微流体平台筛选 | [ |
| 谷氨酸棒杆菌 | ARTP诱变 | 提升L-丝氨酸的生产能力 | 1.2?×?105 | 生物传感器筛选 | [ |
| 莱茵衣藻 | 甲磺酸乙酯化学诱变 | 增加细胞生长和脂质积累 | 2×105 | 液滴微流体平台筛选 | [ |
| 大肠杆菌 | ARTP诱变 | 提高菌株L-赖氨酸的生产能力 | 107 | 生物传感器筛选 | [ |
| 大肠杆菌 | ARTP诱变 | 提高菌株苏氨酸的生产能力 | 2 × 107 | 生物传感器筛选 | [ |
表1 菌种进化工程产生的突变文库及其筛选方法
| 菌株 | 进化手段 | 进化目的 | 突变文库大小 | 筛选方法 | 参考文献 |
|---|---|---|---|---|---|
| 莱茵衣藻 | 基因重组 | 提高细胞脂质积累 | 6×104 | 基于细胞颜色筛选 | [ |
| 酵母 | UV诱变 | 提高菌株α-淀粉酶的分泌能力 | 105 | 液滴微流体平台筛选 | [ |
| 谷氨酸棒杆菌 | ARTP诱变 | 提升L-丝氨酸的生产能力 | 1.2?×?105 | 生物传感器筛选 | [ |
| 莱茵衣藻 | 甲磺酸乙酯化学诱变 | 增加细胞生长和脂质积累 | 2×105 | 液滴微流体平台筛选 | [ |
| 大肠杆菌 | ARTP诱变 | 提高菌株L-赖氨酸的生产能力 | 107 | 生物传感器筛选 | [ |
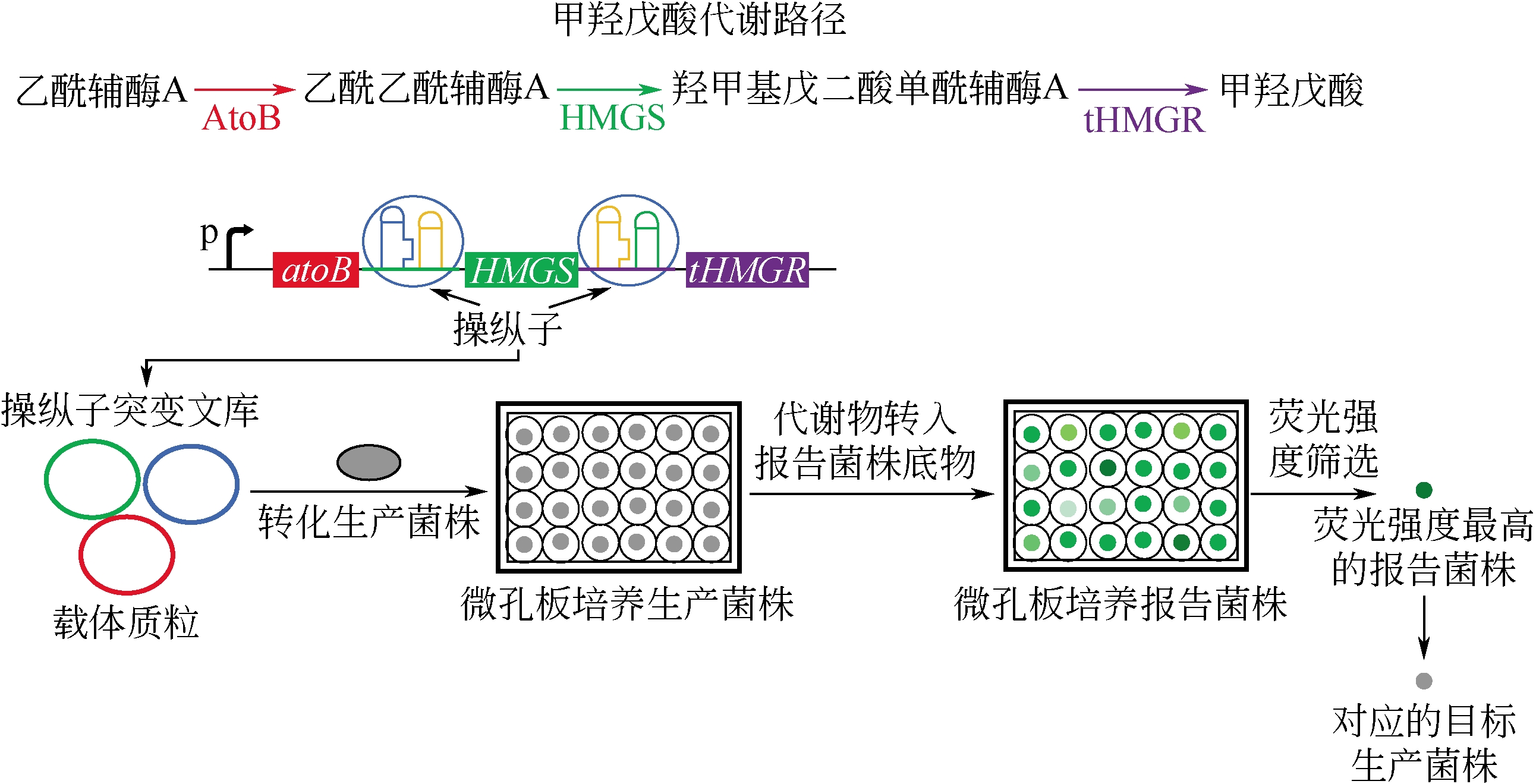
| 大肠杆菌 | ARTP诱变 | 提高菌株苏氨酸的生产能力 | 2 × 107 | 生物传感器筛选 | [ |
| 宿主细胞 | 响应物质 | 生物传感器筛选结果或作用 | 参考文献 |
|---|---|---|---|
| 谷氨酸棒杆菌 | L-缬氨酸 | 目标突变体L-缬氨酸的产量提升了25%,副产物减少了3~4倍 | [ |
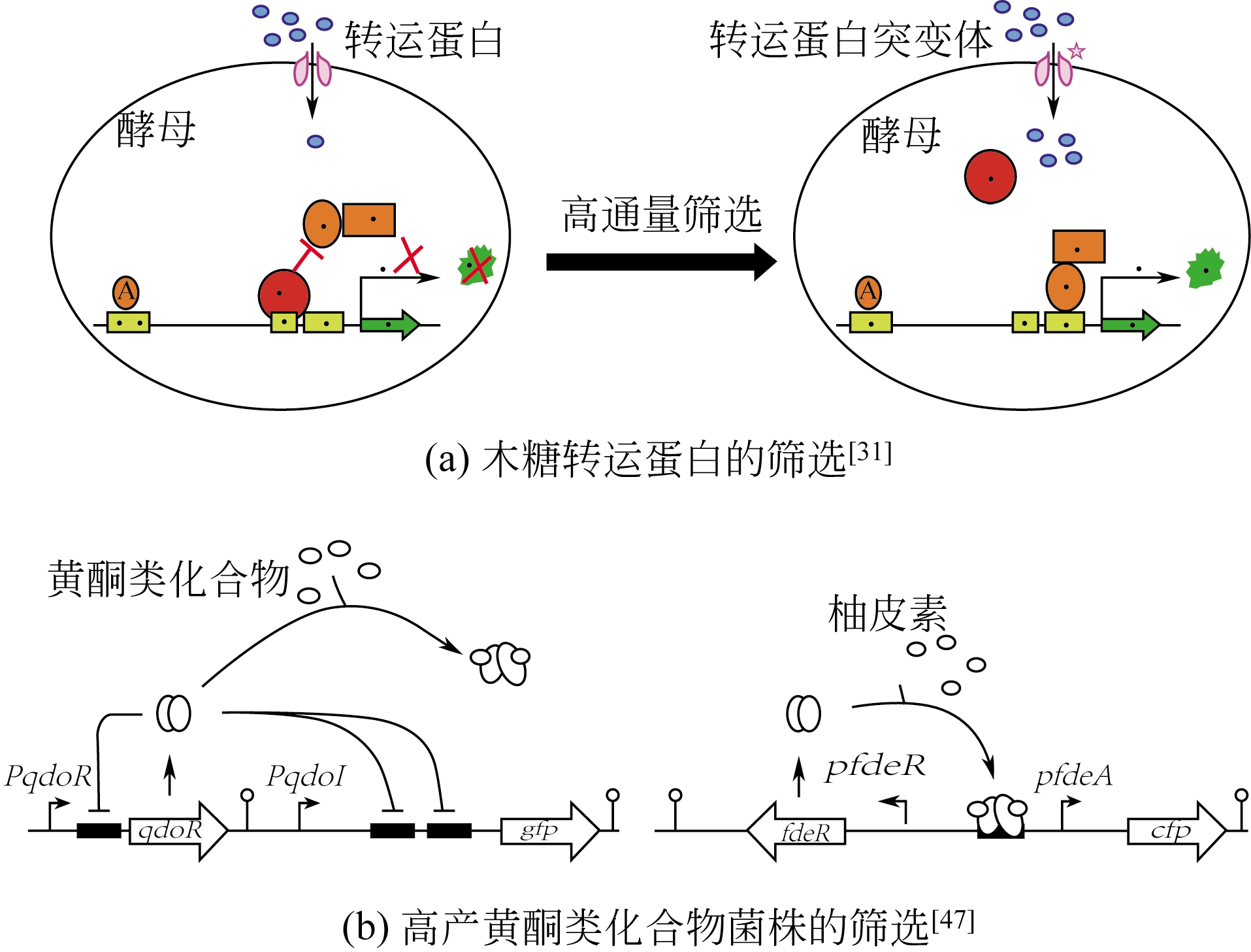
| 酿酒酵母 | 木糖 | 开发了一套木糖转运蛋白高通量筛选方法,并获得了一种优良的突变体,其木糖转运能力提高了6.5倍 | [ |
| 大肠杆菌 | 酪氨酸 | 目标突变体酪氨酸产量提升5倍 | [ |
| 大肠杆菌 | 1-丁醇 | 目标突变体1-丁醇产量提升35% | [ |
| 大肠杆菌 | 葡萄糖二酸和柚皮素 | 目标突变体葡萄糖二酸产量提升22倍,柚皮素产量提升36倍 | [ |
| 大肠杆菌 | 乙醇脱氢酶 | 乙醇脱氢酶对底物的特异性地得到提高 | [ |
| 大肠杆菌 | 三乙酸内酯 | 成功突变天然转录因子 AraC特异性响应细胞内三乙酸内酯 | [ |
| 大肠杆菌 | 甲羟戊酸 | 成功突变天然转录因子 AraC特异性响应细胞内甲羟戊酸 | [ |
| 大肠杆菌 | D-阿拉伯糖 | 成功突变天然转录因子 AraC特异性响应细胞内D-阿拉伯糖信号 | [ |
| 大肠杆菌 | 四氢嘧啶 | 成功突变天然转录因子 AraC特异性响应细胞内四氢嘧啶,并成功筛选到高产四氢嘧啶的目标突变体 | [ |
| 酿酒酵母 | 丙二酰辅酶A | 目标突变体丙二酰辅酶A表达提高,下游产物3-羟基丙酸的产量提升了120% | [ |
表2 基于转录因子的高通量筛选应用
| 宿主细胞 | 响应物质 | 生物传感器筛选结果或作用 | 参考文献 |
|---|---|---|---|
| 谷氨酸棒杆菌 | L-缬氨酸 | 目标突变体L-缬氨酸的产量提升了25%,副产物减少了3~4倍 | [ |
| 酿酒酵母 | 木糖 | 开发了一套木糖转运蛋白高通量筛选方法,并获得了一种优良的突变体,其木糖转运能力提高了6.5倍 | [ |
| 大肠杆菌 | 酪氨酸 | 目标突变体酪氨酸产量提升5倍 | [ |
| 大肠杆菌 | 1-丁醇 | 目标突变体1-丁醇产量提升35% | [ |
| 大肠杆菌 | 葡萄糖二酸和柚皮素 | 目标突变体葡萄糖二酸产量提升22倍,柚皮素产量提升36倍 | [ |
| 大肠杆菌 | 乙醇脱氢酶 | 乙醇脱氢酶对底物的特异性地得到提高 | [ |
| 大肠杆菌 | 三乙酸内酯 | 成功突变天然转录因子 AraC特异性响应细胞内三乙酸内酯 | [ |
| 大肠杆菌 | 甲羟戊酸 | 成功突变天然转录因子 AraC特异性响应细胞内甲羟戊酸 | [ |
| 大肠杆菌 | D-阿拉伯糖 | 成功突变天然转录因子 AraC特异性响应细胞内D-阿拉伯糖信号 | [ |
| 大肠杆菌 | 四氢嘧啶 | 成功突变天然转录因子 AraC特异性响应细胞内四氢嘧啶,并成功筛选到高产四氢嘧啶的目标突变体 | [ |
| 酿酒酵母 | 丙二酰辅酶A | 目标突变体丙二酰辅酶A表达提高,下游产物3-羟基丙酸的产量提升了120% | [ |
| 宿主细胞 | 响应物质 | 生物传感器筛选结果或作用 | 参考文献 |
|---|---|---|---|
| 大肠杆菌 | 茶碱 | 鉴定出了增加茶碱生产的最佳生物元件组合 | [ |
| 大肠杆菌 | 氰尿二酰胺 | 改造设计正交核糖体开关,不再响应细胞内天然配体分子,而特异性响应非天然小分子氰尿二酰胺 | [ |
| 酿酒酵母 | 6-磷酸葡糖胺 | 能够有效分离高产n-乙酰氨基葡糖的突变体 | [ |
| 苜蓿根瘤菌 | VB12 | 获得了一株优良的阳性突变体,VB12产量比野生型提高了21.9% | [ |
表3 基于核糖体开关的高通量筛选应用
| 宿主细胞 | 响应物质 | 生物传感器筛选结果或作用 | 参考文献 |
|---|---|---|---|
| 大肠杆菌 | 茶碱 | 鉴定出了增加茶碱生产的最佳生物元件组合 | [ |
| 大肠杆菌 | 氰尿二酰胺 | 改造设计正交核糖体开关,不再响应细胞内天然配体分子,而特异性响应非天然小分子氰尿二酰胺 | [ |
| 酿酒酵母 | 6-磷酸葡糖胺 | 能够有效分离高产n-乙酰氨基葡糖的突变体 | [ |
| 苜蓿根瘤菌 | VB12 | 获得了一株优良的阳性突变体,VB12产量比野生型提高了21.9% | [ |
| 宿主细胞 | 响应物质 | 生物传感器筛选结果或作用 | 参考文献 |
|---|---|---|---|
| 大肠杆菌 | 6-磷酸海藻糖 | 实时监测细胞内的6-磷酸 海藻糖浓度 | [ |
| 大肠杆菌和 酵母 | 蛋氨酸 | 细胞内蛋氨酸监测 | [ |
| 大肠杆菌 | 亮氨酸 | 细胞内亮氨酸监测 | [ |
| 大肠杆菌 | 抗生素 | 建立了高通量筛选潜在 抗生素分子的方法 | [ |
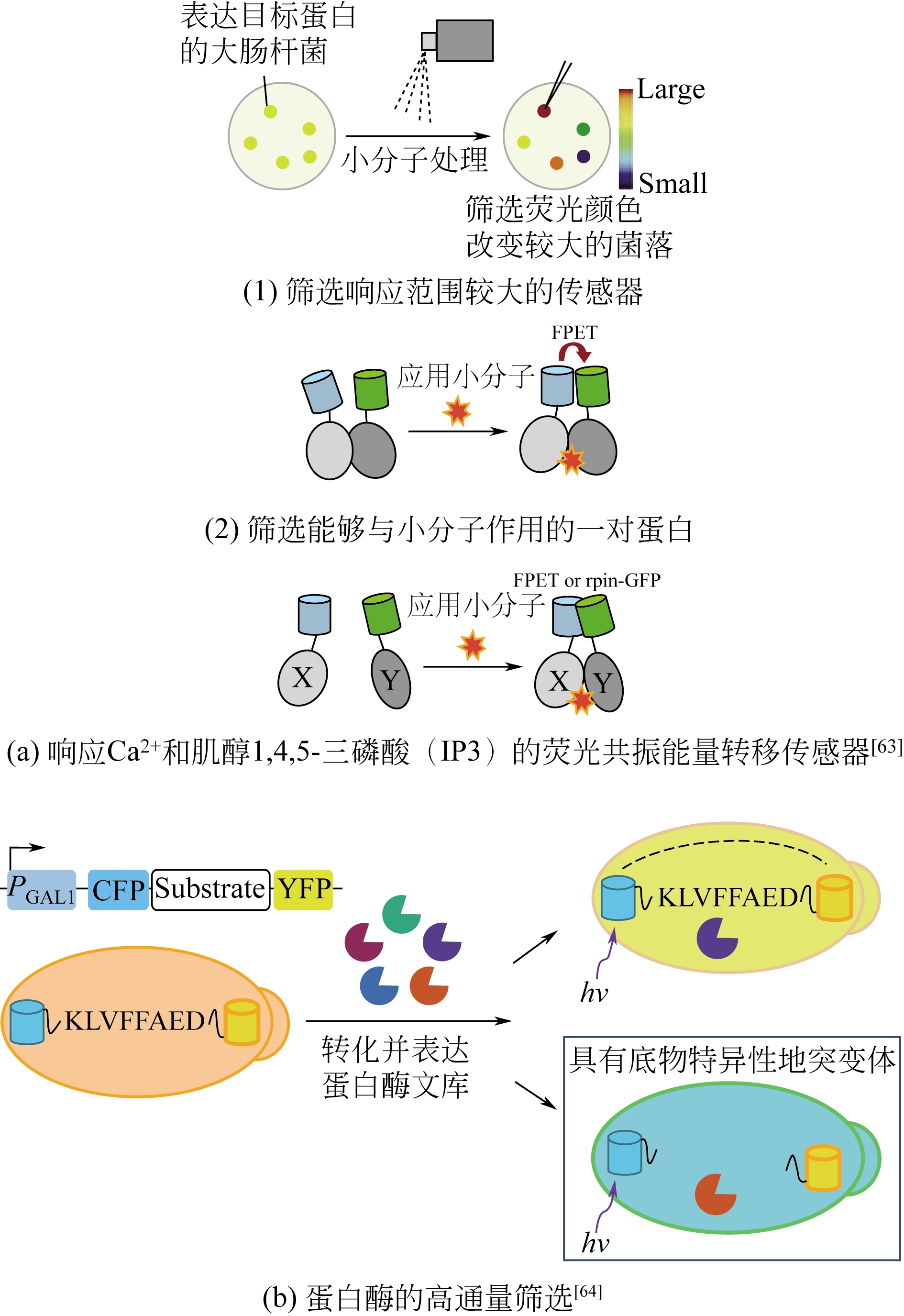
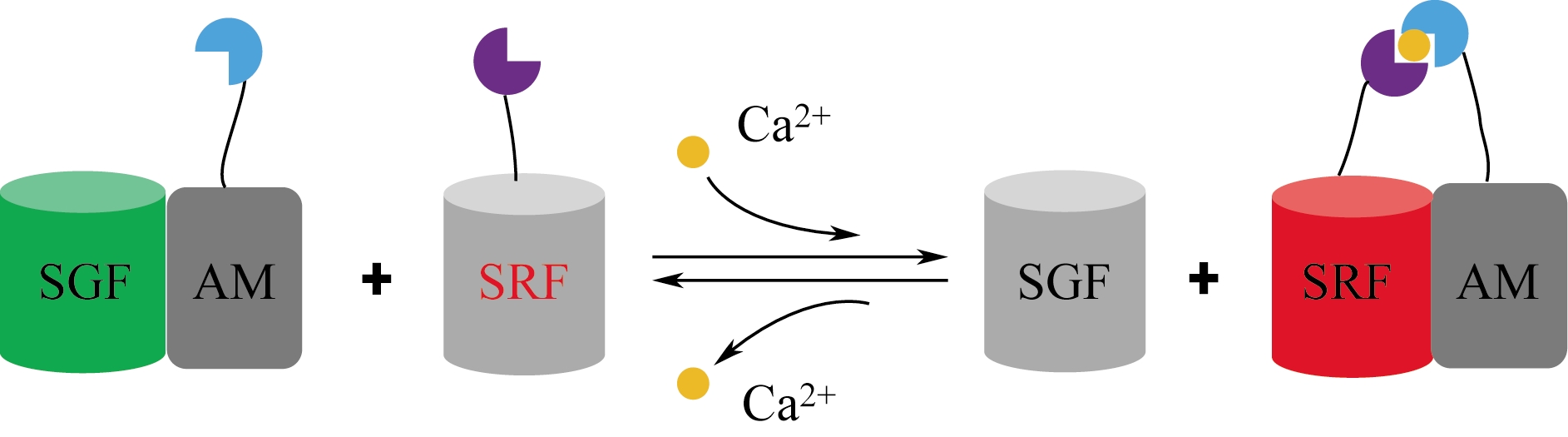
| 大肠杆菌 | 肌醇1,4,5-三磷酸 | 建立了高通量筛选小分子 肌醇1,4,5-三磷酸的方法 | [ |
| 酿酒酵母 | 蛋白酶 | 成功筛选出一种对底物选择性提高30倍的突变体 | [ |
表4 基于荧光共振能量转移的高通量筛选应用
| 宿主细胞 | 响应物质 | 生物传感器筛选结果或作用 | 参考文献 |
|---|---|---|---|
| 大肠杆菌 | 6-磷酸海藻糖 | 实时监测细胞内的6-磷酸 海藻糖浓度 | [ |
| 大肠杆菌和 酵母 | 蛋氨酸 | 细胞内蛋氨酸监测 | [ |
| 大肠杆菌 | 亮氨酸 | 细胞内亮氨酸监测 | [ |
| 大肠杆菌 | 抗生素 | 建立了高通量筛选潜在 抗生素分子的方法 | [ |
| 大肠杆菌 | 肌醇1,4,5-三磷酸 | 建立了高通量筛选小分子 肌醇1,4,5-三磷酸的方法 | [ |
| 酿酒酵母 | 蛋白酶 | 成功筛选出一种对底物选择性提高30倍的突变体 | [ |
| 1 | FERREIRO A , CROOK N , GASPARRINI A J , et al . Multiscale evolutionary dynamics of host-associated microbiomes [J]. Cell, 2018, 172(6): 1216-1227. |
| 2 | WOOLSTON B M , EDGAR S , STEPHANOPOULOS G . Metabolic engineering: past and future[J]. Annual Review of Chemical and Biomolecular Engineering, 2013, 4: 259-288. |
| 3 | TURANLI-YILDIZ B , BENBADIS L , ALKIM C , et al . In vivo evolutionary engineering for ethanol-tolerance of Saccharomyces cerevisiae haploid cells triggers diploidization[J]. Journal of Bioscience and Bioengineering, 2017, 124(3): 309-318. |
| 4 | ZENG W Y , TANG Y Q , GOU M , et al . Comparative transcriptomes reveal novel evolutionary strategies adopted by Saccharomyces cerevisiae with improved xylose utilization capability[J]. Applied Microbiology & Biotechnology, 2017, 101(4): 1753-1767. |
| 5 | SHEPELIN D , HANSEN A S L , LENNEN R , et al . Selecting the best: evolutionary engineering of chemical production in microbes[J]. Genes, 2018, 9(5): 249. |
| 6 | DUARTE J M , BARBIER I , SCHAERLI Y . Bacterial microcolonies in gel beads for high-throughput screening of libraries in synthetic biology[J]. ACS Synthetic Biology, 2017, 6(11): 1988-1995. |
| 7 | LIU X , PAINTER R , ENESA K , et al . High-throughput screening of antibiotic-resistant bacteria in picodroplets[J]. Lab on a Chip, 2016, 16(9): 1636-1643. |
| 8 | TERASHIMA M , FREEMAN E S , JINKERSON R E , et al . A fluorescence-activated cell sorting-based strategy for rapid isolation of high-lipid C hlamydomonas mutants[J]. The Plant Journal, 2015, 81(1): 147-159. |
| 9 | SJOSTROM S L , BAI Y , HUANG M , et al . High-throughput screening for industrial enzyme production hosts by droplet microfluidics[J]. Lab on a Chip, 2014, 14(4): 806-813. |
| 10 | ZHANG X , ZHANG X , XU G , et al . Integration of ARTP mutagenesis with biosensor-mediated high-throughput screening to improve l-serine yield in Corynebacterium glutamicum [J]. Applied Microbiology and Biotechnology, 2018, 102(14):5939-5951. |
| 11 | KIM H S, HSU S C, HAN S I , et al . High-throughput droplet microfluidics screening platform for selecting fast-growing and high lipid-producing microalgae from a mutant library[J]. Plant Direct, 2017, 1(3): e00011. |
| 12 | WANG Y , LI Q , ZHENG P , et al . Evolving the L-lysine high-producing strain of Escherichia coli using a newly developed high-throughput screening method[J]. Journal of Industrial Microbiology & Biotechnology, 2016, 43(9): 1227-1235. |
| 13 | LIU Y N , LI Q , ZHENG P , et al . Developing a high-throughput screening method for threonine overproduction based on an artificial promoter[J]. Microbial Cell Factories, 2015, 14(1): 121. |
| 14 | ALPER H , MIYAOKU K , STEPHANOPOULOS G . Construction of lycopene-overproducing E. coli strains by combining systematic and combinatorial gene knockout targets[J]. Nature Biotechnology, 2005, 23(5): 612. |
| 15 | LI J , SHEN J , SUN Z , et al . Discovery of several novel targets that enhance β-carotene production in Saccharomyces cerevisiae [J]. Frontiers in Microbiology, 2017, 8:1116. |
| 16 | ZHOU P , XIE W , LI A , et al . Alleviation of metabolic bottleneck by combinatorial engineering enhanced astaxanthin synthesis in Saccharomyces cerevisiae [J]. Enzyme and Microbial Technology, 2017, 100:28-36. |
| 17 | LIU W , JIANG R . Combinatorial and high-throughput screening approaches for strain engineering[J]. Applied Microbiology and Biotechnology, 2015, 99(5): 2093-2104. |
| 18 | DIETRICH J A , MCKEE A E , KEASLING J D . High-throughput metabolic engineering: advances in small-molecule screening and selection[J]. Annual Review of Biochemistry, 2010, 79:563-590. |
| 19 | SCHALLMEY M , FRUNZKE J , EGGELING L , et al . Looking for the pick of the bunch: high-throughput screening of producing microorganisms with biosensors[J]. Current Opinion in Biotechnology, 2014, 26(26C): 148-154. |
| 20 | UKIBE K , KATSURAGI T , TANI Y , et al . Efficient screening for astaxanthin-overproducing mutants of the yeast Xanthophyllomyces dendrorhous by flow cytometry[J]. FEMS Microbiology Letters, 2008, 286(2): 241-248. |
| 21 | LEE J H, LEE S H, YIM S S, et al . Quantified high-throughput screening of Escherichia coli producing poly(3-hydroxybutyrate) based on FACS[J]. Applied Biochemistry and Biotechnology, 2013, 170(7): 1767-1779. |
| 22 | DELOACHE W C , RUSS Z N , NARCROSS L , et al . An enzyme-coupled biosensor enables (S)-reticuline production in yeast from glucose[J]. Nature Chemical Biology, 2015, 11(7): 465. |
| 23 | HENNING H , LEGGEWIE C , POHL M , et al . Identification of novel benzoylformate decarboxylases by growth selection[J]. Applied and Environmental Microbiology, 2006, 72(12): 7510-7517. |
| 24 | PFLEGER B F , PITERA D J , SMOLKE C D , et al . Combinatorial engineering of intergenic regions in operons tunes expression of multiple genes[J]. Nature Biotechnology, 2006, 24(8): 1027. |
| 25 | BOERSMA Y L , DRÖGE M J , SLOOT A M VAN DER , et al . A novel genetic selection system for improved enantioselectivity of Bacillus subtilis lipase A[J]. Chem. Bio. Chem., 2008, 9(7): 1110-1115. |
| 26 | BOLES E , OREB M . A growth-based screening system for hexose transporters in yeast [M]// Glucose Transport. Springer, 2018: 123-135. |
| 27 | MICHENER J K , THODEY K , LIANG J C , et al . Applications of genetically-encoded biosensors for the construction and control of biosynthetic pathways[J]. Metabolic Engineering, 2012, 14(3): 212-222. |
| 28 | LATCHMAN D S . Transcription factors: an overview[J]. The International Journal of Biochemistry & Cell Biology, 1997, 29(12): 1305-1312. |
| 29 | BINDER S , SCHENDZIELORZ G , BLER N ST , et al . A high-throughput approach to identify genomic variants of bacterial metabolite producers at the single-cell level[J]. Genome Biology, 2012, 13(5): R40. |
| 30 | MAHR R , GÄTGENS C , GÄTGENS J , et al . Biosensor-driven adaptive laboratory evolution of L-valine production in Corynebacterium glutamicum [J]. Metabolic Engineering, 2015, 32:184-194. |
| 31 | WANG M , LI S , ZHAO H . Design and engineering of intracellular-metabolite-sensing/regulation gene circuits in Saccharomyces cerevisiae [J]. Biotechnology and Bioengineering, 2016, 113(1): 206-215. |
| 32 | CHOU H H , KEASLING J D . Programming adaptive control to evolve increased metabolite production[J]. Nature Communications, 2013, 4:2595. |
| 33 | DIETRICH J A , SHIS D L , ALIKHANI A , et al . Transcription factor-based screens and synthetic selections for microbial small-molecule biosynthesis[J]. ACS Synthetic Biology, 2012, 2(1): 47-58. |
| 34 | RAMAN S , ROGERS J K , TAYLOR N D , et al . Evolution-guided optimization of biosynthetic pathways[J]. Proceedings of the National Academy of Sciences, 2014, 111(50): 17803-17808. |
| 35 | SIEDLER S , SCHENDZIELORZ G , BINDER S , et al . So x R as a single-cell biosensor for NADPH-consuming enzymes in Escherichia coli [J]. ACS Synthetic Biology, 2013, 3(1): 41-47. |
| 36 | SIERRO N , MAKITA Y , HOON M DE , et al . DBTBS: a database of transcriptional regulation in Bacillus subtilis containing upstream intergenic conservation information[J]. Nucleic Acids Research, 2007, 36(suppl1): D93-D96. |
| 37 | GAMA-CASTRO S , NEZ-JACINTO V JIM , PERALTA-GIL M , et al . RegulonDB (version 6.0): gene regulation model of Escherichia coli K-12 beyond transcription, active (experimental) annotated promoters and textpresso navigation[J]. Nucleic Acids Research, 2008, 36(suppl1): D120-D124. |
| 38 | MATYS V , KEL-MARGOULIS O V , FRICKE E , et al . TRANSFAC® and its module TRANSCompel®: transcriptional gene regulation in eukaryotes[J]. Nucleic Acids Research, 2006, 34(s1): D108-D110. |
| 39 | KOLCHANOV N A , IGNATIEVA E V , ANANKO E A , et al . Transcription regulatory regions database (TRRD): its status in 2002[J]. Nucleic Acids Research, 2002, 30(1): 312-317. |
| 40 | CIPRIANO M J , NOVICHKOV P N , KAZAKOV A E , et al . RegTransBase——a database of regulatory sequences and interactions based on literature: a resource for investigating transcriptional regulation in prokaryotes[J]. BMC Genomics, 2013, 14(1): 213. |
| 41 | FREI C S , WANG Z , QIAN S , et al . Analysis of amino acid substitutions in AraC variants that respond to triacetic acid lactone[J]. Protein Science, 2016, 25(4): 804-814. |
| 42 | TANG S-Y , QIAN S , AKINTERINWA O , et al . Screening for enhanced triacetic acid lactone production by recombinant Escherichia coli expressing a designed triacetic acid lactone reporter[J]. Journal of the American Chemical Society, 2013, 135(27): 10099-10103. |
| 43 | TANG S Y , CIRINO P C . Design and application of a mevalonate-responsive regulatory protein[J]. Angewandte Chemie, 2011, 123(5): 1116-1118. |
| 44 | TANG S Y , FAZELINIA H , CIRINO P C . AraC regulatory protein mutants with altered effector specificity[J]. Journal of the American Chemical Society, 2008, 130(15): 5267-5271. |
| 45 | CHEN W , ZHANG S , JIANG P , et al . Design of an ectoine-responsive AraC mutant and its application in metabolic engineering of ectoine biosynthesis[J]. Metabolic Engineering, 2015, 30:149-155. |
| 46 | LI S , SI T , WANG M , et al . Development of a synthetic malonyl-CoA sensor in Saccharomyces cerevisiae for intracellular metabolite monitoring and genetic screening[J]. ACS Synthetic Biology, 2015, 4(12): 1308-1315. |
| 47 | SIEDLER S , STAHLHUT S G , MALLA S , et al . Novel biosensors based on flavonoid-responsive transcriptional regulators introduced into Escherichia coli [J]. Metabolic Engineering, 2014, 21:2-8. |
| 48 | BASTET L , TURCOTTE P , WADE J T , et al . Maestro of regulation: riboswitches orchestrate gene expression at the levels of translation, transcription and mRNA decay[J]. RNA Biology, 2018, 15(6):679-682. |
| 49 | ECKDAHL T T , CAMPBELL A M , HEYER L J , et al . Programmed evolution for optimization of orthogonal metabolic output in bacteria[J]. PLoS One, 2015, 10(2): e0118322. |
| 50 | DIXON N , DUNCAN J N , GEERLINGS T , et al . Reengineering orthogonally selective riboswitches[J]. Proceedings of the National Academy of Sciences, 2010, 107(7): 2830-2835. |
| 51 | LEE S W, OH M K . A synthetic suicide riboswitch for the high-throughput screening of metabolite production in Saccharomyces cerevisiae [J]. Metabolic Engineering, 2015, 28: 143-150. |
| 52 | CAI Y , XIA M , DONG H , et al . Engineering a vitamin B12 high-throughput screening system by riboswitch sensor in Sinorhizobium meliloti [J]. BMC Biotechnology, 2018, 18(1): 27. |
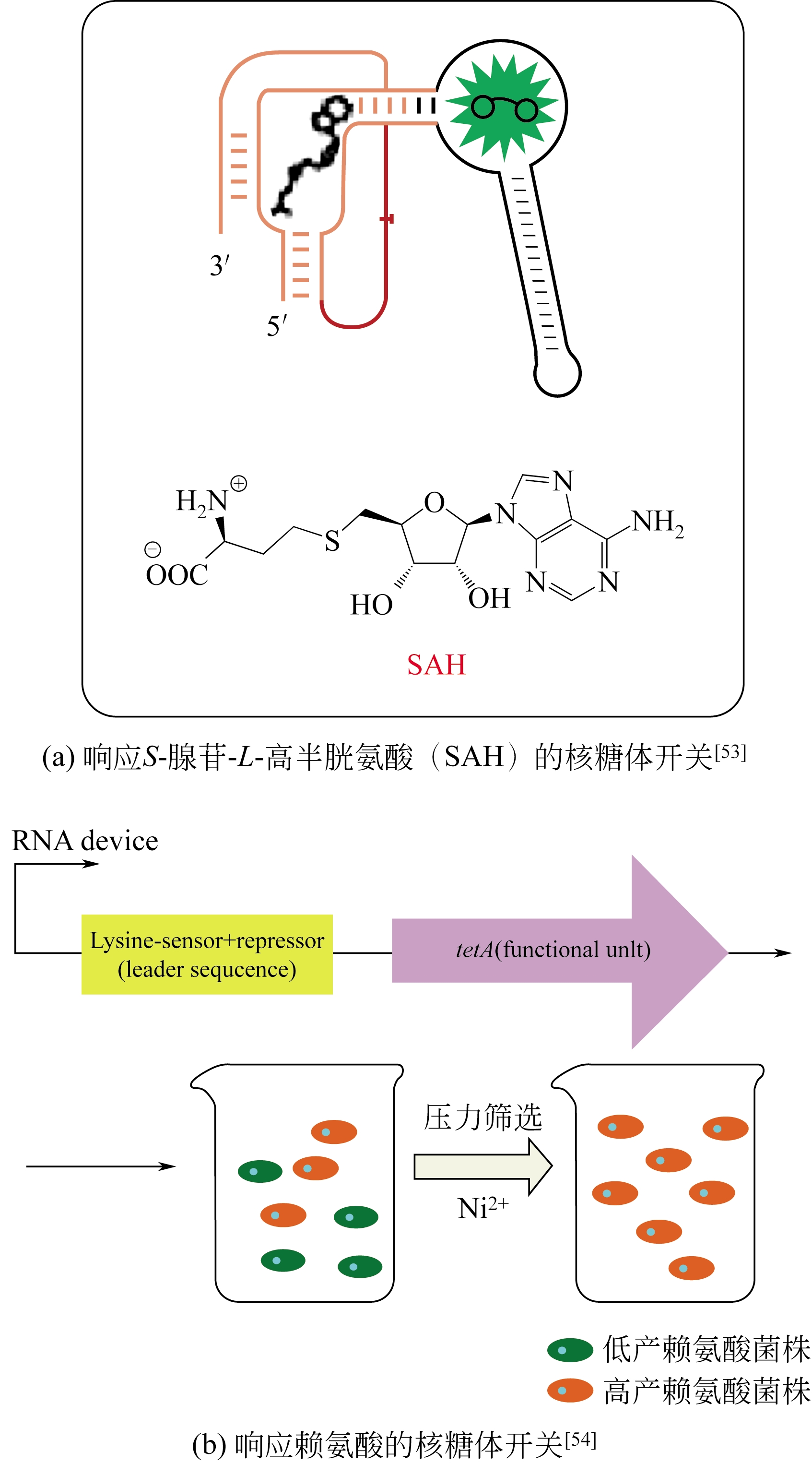
| 53 | SU Y , HICKEY S F , KEYSER S G , et al . In vitro and in vivo enzyme activity screening via RNA-based fluorescent biosensors for S-adenosyl-L-homocysteine (SAH)[J]. Journal of the American Chemical Society, 2016, 138(22): 7040-7047. |
| 54 | WANG J , GAO D , YU X , et al . Evolution of a chimeric aspartate kinase for L-lysine production using a synthetic RNA device[J]. Applied Microbiology and Biotechnology, 2015, 99(20): 8527-8536. |
| 55 | TRAUSCH J J , CERES P , REYES F E , et al . The structure of a tetrahydrofolate-sensing riboswitch reveals two ligand binding sites in a single aptamer[J]. Structure, 2011, 19(10): 1413-1423. |
| 56 | WACHSMUTH M , FINDEI S , WEISSHEIMER N , et al . De novo design of a synthetic riboswitch that regulates transcription termination[J]. Nucleic Acids Research, 2012, 41(4): 2541-2551. |
| 57 | TOPP S , REYNOSO C M , SEELIGER J C , et al . Synthetic riboswitches that induce gene expression in diverse bacterial species[J]. Applied and Environmental Microbiology, 2010, 76(23): 7881-7884. |
| 58 | ESPAH BORUJENI A , MISHLER D M , WANG J , et al . Automated physics-based design of synthetic riboswitches from diverse RNA aptamers[J]. Nucleic Acids Research, 2015, 44(1): 1-13. |
| 59 | PEROZA E A , EWALD J C , PARAKKAL G , et al . A genetically encoded Förster resonance energy transfer sensor for monitoring in vivo trehalose-6-phosphate dynamics[J]. Analytical Biochemistry, 2015, 474:1-7. |
| 60 | MOHSIN M , AHMAD A . Genetically-encoded nanosensor for quantitative monitoring of methionine in bacterial and yeast cells[J]. Biosensors and Bioelectronics, 2014, 59:358-364. |
| 61 | MOHSIN M , ABDIN M , NISCHAL L , et al . Genetically encoded FRET-based nanosensor for in vivo measurement of leucine[J]. Biosensors and Bioelectronics, 2013, 50:72-77. |
| 62 | BHATT R , CHUDAEV M , MANDECKI W , et al . Engineered EF-Tu and tRNA-based FRET screening assay to find inhibitors of protein synthesis in bacteria[J]. Assay and Drug Development Technologies, 2018, 16(4):212-221. |
| 63 | MIYAMOTO A , SUGIURA K , MIKOSHIBA K . Development of a convenient and supersensitive high-throughput screening system for genetically encoded fluorescent probes of small molecules using a confocal microscope[J]. Cell Calcium, 2017, 61: 1-9. |
| 64 | GUERRERO J L , O’MALLEY M A , DAUGHERTY P S . Intracellular FRET-based screen for redesigning the specificity of secreted proteases[J]. ACS Chemical Biology, 2016, 11(4): 961-970. |
| 65 | BEHJOUSIAR A , KONTORAVDI C , POLIZZI K M . In situ monitoring of intracellular glucose and glutamine in CHO cell culture[J]. PLoS One, 2012, 7(4): e34512. |
| 66 | WANG J , WEI J , SU S , et al . Novel fluorescence resonance energy transfer optical sensors for vitamin B12 detection using thermally reduced carbon dots[J]. New Journal of Chemistry, 2015, 39(1): 501-507. |
| 67 | NGUYEN T-T T , TAWFIK S M , ZAYAKHUU G , et al . Highly selective and sensitive optosensing of glutathione based on fluorescence resonance energy transfer of upconversion nanoparticles coated with a Rhodamine B derivative[J]. Arabian Journal of Chemistry, 2018, . |
| 68 | DING Y , LI J , ENTERINA J R , et al . Ratiometric biosensors based on dimerization-dependent fluorescent protein exchange[J]. Nature Methods, 2015, 12(3): 195. |
| 69 | SZITA N , POLIZZI K , JACCARD N , et al . Microfluidic approaches for systems and synthetic biology[J]. Current Opinion in Biotechnology, 2010, 21(4): 517-523. |
| 70 | AGRESTI J J , ANTIPOV E , ABATE A R , et al . Ultrahigh-throughput screening in drop-based microfluidics for directed evolution[J]. Proceedings of the National Academy of Sciences, 2010, 107(9):4004-4009. |
| 71 | MA F, CHUNG M T , YAO Y , et al . Efficient molecular evolution to generate enantioselective enzymes using a dual-channel microfluidic droplet screening platform[J]. Nature communications, 2018, 9(1): 1030. |
| 72 | HUANG M , BAI Y , SJOSTROM S L , et al . Microfluidic screening and whole-genome sequencing identifies mutations associated with improved protein secretion by yeast[J]. Proceedings of the National Academy of Sciences, 2015, 112(34): E4689-E4696. |
| 73 | WANG B L , GHADERI A , ZHOU H , et al . Microfluidic high-throughput culturing of single cells for selection based on extracellular metabolite production or consumption[J]. Nature Biotechnology, 2014, 32(5): 473. |
| 74 | MACAULAY I C , PONTING C P , VOET T . Single-cell multiomics: multiple measurements from single cells[J]. Trends in Genetics, 2017, 33(2): 155-168. |
| 75 | ABATE A R , HUNG T , SPERLING R A , et al . DNA sequence analysis with droplet-based microfluidics[J]. Lab on a Chip, 2013, 13(24): 4864-4869. |
| 76 | TREUTLEIN B , BROWNFIELD D G , WU A R , et al . Reconstructing lineage hierarchies of the distal lung epithelium using single-cell RNA-seq[J]. Nature, 2014, 509(7500): 371. |
| 77 | MOON H S , JE K, MIN J W , et al . Inertial-ordering-assisted droplet microfluidics for high-throughput single-cell RNA-sequencing[J]. Lab on a Chip, 2018, 18(5): 775-784. |
| 78 | VALIHRACH L , ANDROVIC P , KUBISTA M . Platforms for single-cell collection and analysis[J]. International Journal of Molecular Sciences, 2018, 19(3): 807. |
| 79 | WANG J , SONG Y . Single cell sequencing: a distinct new field[J]. Clinical and Translational Medicine, 2017, 6(1): 10. |
| 80 | FENG Y , ZHANG Y , YING C , et al . Nanopore-based fourth-generation DNA sequencing technology[J]. Genomics, Proteomics & Bioinformatics, 2015, 13(1): 4-16. |
| 81 | SCHREIBER J , WESCOE Z L , ABU-SHUMAYS R , et al . Error rates for nanopore discrimination among cytosine, methylcytosine, and hydroxymethylcytosine along individual DNA strands[J]. Proceedings of the National Academy of Sciences, 2013, 201310615. |
| 82 | GARALDE D R , SNELL E A , JACHIMOWICZ D , et al . Highly parallel direct RNA sequencing on an array of nanopores[J]. Nature Methods, 2018, 15(3): 201. |
| 83 | VILFAN I D , TSAI Y C , CLARK T A , et al . Analysis of RNA base modification and structural rearrangement by single-molecule real-time detection of reverse transcription[J]. Journal of Nanobiotechnology, 2013, 11(1): 8. |
| 84 | ROSEN C B , RODRIGUEZ-LARREA D , BAYLEY H . Single-molecule site-specific detection of protein phosphorylation with a nanopore[J]. Nature Biotechnology, 2014, 32(2): 179. |
| 85 | XIE Z X , LI B Z , MITCHELL L A , et al . “Perfect” designer chromosome V and behavior of a ring derivative[J]. Science, 2017, 355(6329): eaaf4704. |
| 86 | WU Y , LI B-Z , ZHAO M , et al . Bug mapping and fitness testing of chemically synthesized chromosome X[J]. Science, 2017, 355(6329): eaaf4706. |
| 87 | JIA B , WU Y , LI B-Z , et al . Precise control of SCRaMbLE in synthetic haploid and diploid yeast[J]. Nature Communications, 2018, 9(1): 1933. |
| [1] | 孙 艳,杨海麟,王 武. 建立高通量筛选耐热胆固醇氧化酶的方法 [J]. 化工进展, 2011, 30(3): 612-. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||