Chemical Industry and Engineering Progress ›› 2024, Vol. 43 ›› Issue (S1): 533-544.DOI: 10.16085/j.issn.1000-6613.2024-1023
• Resources and environmental engineering • Previous Articles Next Articles
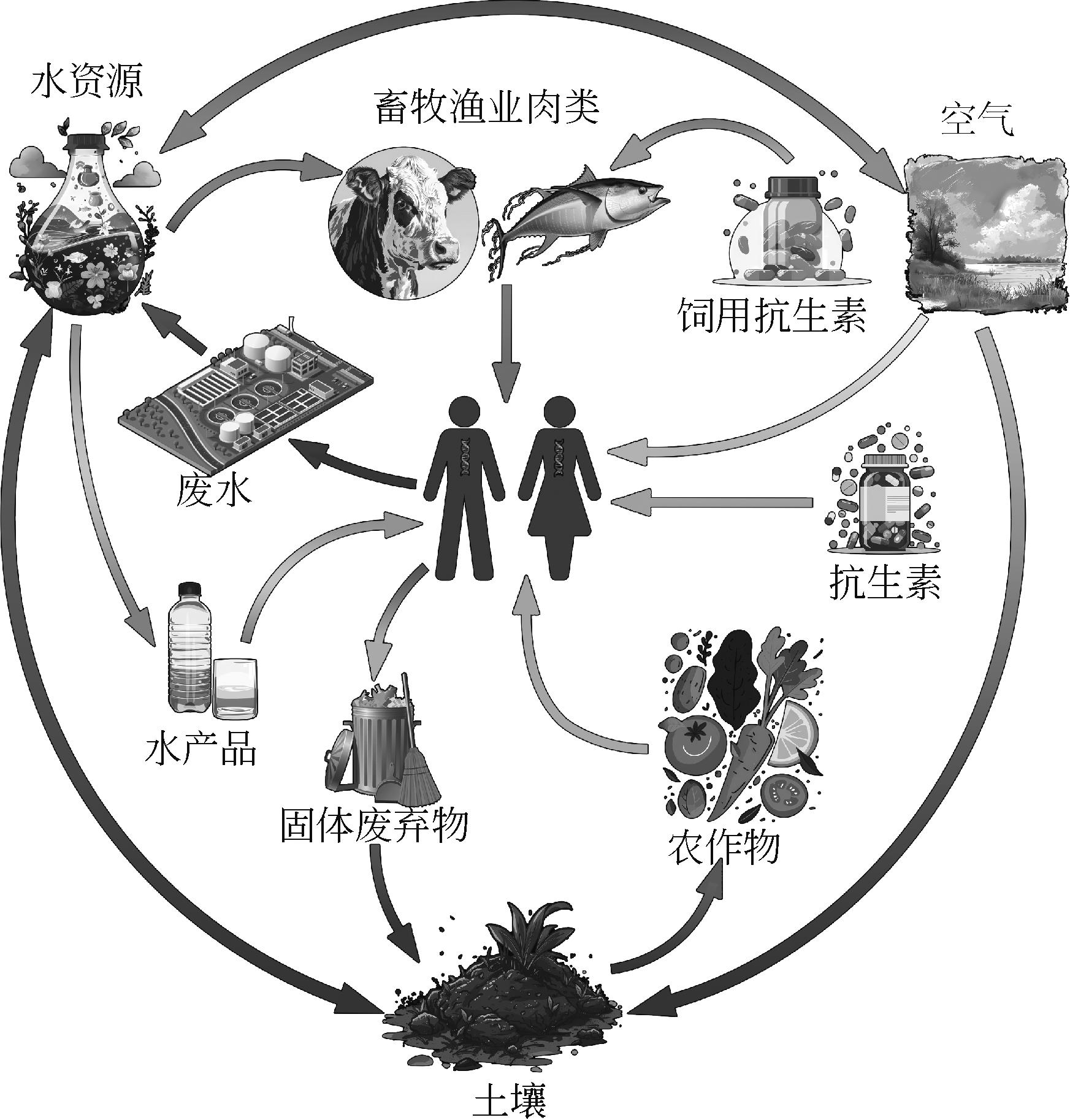
Progress in the study of antibiotic resistance genes in the aquatic environment
HE Zihan1( ), LI Wenxuan2, LI Yanyu1, WANG Xuechao1, YANG Shirong1, XIE Huina1, LI Jie1(
), LI Wenxuan2, LI Yanyu1, WANG Xuechao1, YANG Shirong1, XIE Huina1, LI Jie1( )
)
- 1.School of Environmental and Municipal Engineering, Lanzhou Jiaotong University, Lanzhou 730070, Gansu, China
2.State Key Laboratory of Environmental Criteria and Risk Assessment, State Environmental Protection Key Laboratory of Ecological Effect and Risk Asses sment of Chemicals, Chinese Research Academy of Environmental Sciences, Beijing 100012, China
-
Received:2024-06-25Revised:2024-08-02Online:2024-12-06Published:2024-11-20 -
Contact:LI Jie
水环境中抗生素抗性基因研究进展
何子晗1( ), 李文譞2, 李彦宇1, 王雪超1, 杨始蓉1, 谢慧娜1, 李杰1(
), 李文譞2, 李彦宇1, 王雪超1, 杨始蓉1, 谢慧娜1, 李杰1( )
)
- 1.兰州交通大学环境工程学院,甘肃 兰州 730070
2.中国环境科学研究院国家环境保护化学品生态效应 与风险评估重点实验室,北京 100012
-
通讯作者:李杰 -
作者简介:何子晗(1998—),男,硕士研究生,研究方向为水污染控制。E-mail:497679481@qq.com。 -
基金资助:国家自然科学基金(51768032);甘肃省青年科技基金(23JRRA888)
CLC Number:
Cite this article
HE Zihan, LI Wenxuan, LI Yanyu, WANG Xuechao, YANG Shirong, XIE Huina, LI Jie. Progress in the study of antibiotic resistance genes in the aquatic environment[J]. Chemical Industry and Engineering Progress, 2024, 43(S1): 533-544.
何子晗, 李文譞, 李彦宇, 王雪超, 杨始蓉, 谢慧娜, 李杰. 水环境中抗生素抗性基因研究进展[J]. 化工进展, 2024, 43(S1): 533-544.
share this article
Add to citation manager EndNote|Ris|BibTeX
URL: https://hgjz.cip.com.cn/EN/10.16085/j.issn.1000-6613.2024-1023
| 类型 | 地点 | 情况 | 参考文献 |
|---|---|---|---|
| 海洋 | 厦门市近岸海域 | 共检测出187种ARGs,绝对丰度为1.29×1010 copies/L | [ |
| 秦皇岛昌黎县水产养殖海域 | 共检测到5个β-内酰胺类ARGs、4个氨基糖苷类ARGs | [ | |
| 秦皇岛市五个典型滨海浴场及其邻近河口 | 共检测出159种不同的ARGs,滨海浴场的水体样本中ARGs的相对丰度在0.094~2.5 copies/16S rRNA gene之间,接近甚至高于邻近河口样本 | [ | |
| 北太平洋环流的塑料 | 多重耐药类抗性基因和多金属抗性基因是被检出的主要类别,塑料微生物 群落中ARGs丰度在7.07×10-4~1.21×10-2copies/16S rRNA之间 | [ | |
| 中国黄海四十里湾 | 高频检出10种ARGs亚型,分别为磺胺类、四环素类、喹诺酮类和大环内 酯类。四十里湾ARGs的绝对丰度为100~107copies/mL | [ | |
| 湖泊及水库 | 武汉湖泊(黄家湖、青菱湖、严西湖和涨渡湖)及西藏湖泊(羊卓雍错、空母错、珍错和错那) | 湖泊中共检出21种ARGs和313种ARGs亚型,主要为杆菌肽类、多重耐药 类。武汉湖泊中多黏菌素类和磺胺类丰度分别是西藏的3.5倍和6倍,并且沉 积物中的ARGs显著高于湖泊水体 | [ |
| 上海市金泽水库 | 共检测出283种ARGs,绝对丰度在n.d.-7.97×109 copies/L,其中丰度最高的 为多重抗药性,其次是氨基糖苷类,最低的是氯霉素类 | [ | |
| 江苏省淀山湖-澄湖-太浦河-金泽水库 | ARGs和移动基因元件(mobile genetic elements, MGEs)相对丰度随季节变 化。湖泊、河流及水库中共有54.5%的ARGs亚型 | [ | |
| 中国15个主要湖泊 | 畜牧业对湖泊中的ARGs的分布影响显著。长江中下游平原湖泊ARGs比青 藏高原的更丰富,湖泊沉积物中的四环素类ARGs更丰富 | [ | |
| 河流 | 滏阳河邯郸段 | ARGs绝对丰度在0~2.24×105 copies/L(水)和0~3.57×109 copies/kg(沉积 物),且出市断面丰度为入市断面的19倍 | [ |
| 贵州省赤水河流域 | 共检出9种ARGs,ARGs绝对丰度在1.53×106~1.41×108 copies/L之间(枯水 期)和3.62×104~1.26×108 copies/L之间(丰水期) | [ | |
| 印度亚穆纳河 | 共检出40种ARGs,亚穆纳河种的微生物群落拥有大量的ARGs,显示出对 多种抗生素的耐药性 | [ | |
| 长江(江苏段) | 定量分析了5种ARGs,其绝对丰度在2.81×102~1.31×1010 copies/mL之间, 主要的ARGs为磺酰胺类 | [ | |
| 深圳市茂州河道恢复前后(2018年和2020年) | ARGs和MGEs总丰度为1.41×106 copies/L(2018年)和1.41×105 copies/L (2020年)。河道恢复后,ARGs分布更均匀,并且与微生物群落之间的相关 性增强 | [ |
| 类型 | 地点 | 情况 | 参考文献 |
|---|---|---|---|
| 海洋 | 厦门市近岸海域 | 共检测出187种ARGs,绝对丰度为1.29×1010 copies/L | [ |
| 秦皇岛昌黎县水产养殖海域 | 共检测到5个β-内酰胺类ARGs、4个氨基糖苷类ARGs | [ | |
| 秦皇岛市五个典型滨海浴场及其邻近河口 | 共检测出159种不同的ARGs,滨海浴场的水体样本中ARGs的相对丰度在0.094~2.5 copies/16S rRNA gene之间,接近甚至高于邻近河口样本 | [ | |
| 北太平洋环流的塑料 | 多重耐药类抗性基因和多金属抗性基因是被检出的主要类别,塑料微生物 群落中ARGs丰度在7.07×10-4~1.21×10-2copies/16S rRNA之间 | [ | |
| 中国黄海四十里湾 | 高频检出10种ARGs亚型,分别为磺胺类、四环素类、喹诺酮类和大环内 酯类。四十里湾ARGs的绝对丰度为100~107copies/mL | [ | |
| 湖泊及水库 | 武汉湖泊(黄家湖、青菱湖、严西湖和涨渡湖)及西藏湖泊(羊卓雍错、空母错、珍错和错那) | 湖泊中共检出21种ARGs和313种ARGs亚型,主要为杆菌肽类、多重耐药 类。武汉湖泊中多黏菌素类和磺胺类丰度分别是西藏的3.5倍和6倍,并且沉 积物中的ARGs显著高于湖泊水体 | [ |
| 上海市金泽水库 | 共检测出283种ARGs,绝对丰度在n.d.-7.97×109 copies/L,其中丰度最高的 为多重抗药性,其次是氨基糖苷类,最低的是氯霉素类 | [ | |
| 江苏省淀山湖-澄湖-太浦河-金泽水库 | ARGs和移动基因元件(mobile genetic elements, MGEs)相对丰度随季节变 化。湖泊、河流及水库中共有54.5%的ARGs亚型 | [ | |
| 中国15个主要湖泊 | 畜牧业对湖泊中的ARGs的分布影响显著。长江中下游平原湖泊ARGs比青 藏高原的更丰富,湖泊沉积物中的四环素类ARGs更丰富 | [ | |
| 河流 | 滏阳河邯郸段 | ARGs绝对丰度在0~2.24×105 copies/L(水)和0~3.57×109 copies/kg(沉积 物),且出市断面丰度为入市断面的19倍 | [ |
| 贵州省赤水河流域 | 共检出9种ARGs,ARGs绝对丰度在1.53×106~1.41×108 copies/L之间(枯水 期)和3.62×104~1.26×108 copies/L之间(丰水期) | [ | |
| 印度亚穆纳河 | 共检出40种ARGs,亚穆纳河种的微生物群落拥有大量的ARGs,显示出对 多种抗生素的耐药性 | [ | |
| 长江(江苏段) | 定量分析了5种ARGs,其绝对丰度在2.81×102~1.31×1010 copies/mL之间, 主要的ARGs为磺酰胺类 | [ | |
| 深圳市茂州河道恢复前后(2018年和2020年) | ARGs和MGEs总丰度为1.41×106 copies/L(2018年)和1.41×105 copies/L (2020年)。河道恢复后,ARGs分布更均匀,并且与微生物群落之间的相关 性增强 | [ |
| 环境 | ARGs | 亚型 |
|---|---|---|
| 污水处理厂 | β-内酰胺类 | bla,L1 beta-lactamase,OXA-37,OXA-119,PEDO-1,mecA |
| 磺胺类 | sul,leuO,sul1,sul2,dhfr1 | |
| 四环素类 | tetA,tetB,tetE,tetG,tetH,tetS,tetT,tetX, | |
| 氨基糖苷类 | aadA,aacA4,aadE,strB | |
| 大环内酯类 | ermB,ermC,erm43,mphB,ErmS,ereA,mphG,mefC | |
| 污水管道及管道壁膜生物反应器 | 氨基糖苷类 | AAC(6’)-Iad,aadA13 |
| 磺胺类 | sul1,sul2 | |
| 四环素类 | tetA,tetG | |
| 万古霉素类 | vanA,vanX | |
| β-内酰胺类 | bla,amp | |
| 大环内酯类 | erm |
| 环境 | ARGs | 亚型 |
|---|---|---|
| 污水处理厂 | β-内酰胺类 | bla,L1 beta-lactamase,OXA-37,OXA-119,PEDO-1,mecA |
| 磺胺类 | sul,leuO,sul1,sul2,dhfr1 | |
| 四环素类 | tetA,tetB,tetE,tetG,tetH,tetS,tetT,tetX, | |
| 氨基糖苷类 | aadA,aacA4,aadE,strB | |
| 大环内酯类 | ermB,ermC,erm43,mphB,ErmS,ereA,mphG,mefC | |
| 污水管道及管道壁膜生物反应器 | 氨基糖苷类 | AAC(6’)-Iad,aadA13 |
| 磺胺类 | sul1,sul2 | |
| 四环素类 | tetA,tetG | |
| 万古霉素类 | vanA,vanX | |
| β-内酰胺类 | bla,amp | |
| 大环内酯类 | erm |
| 工艺 | 特点 | 参考文献 |
|---|---|---|
| 生物处理 | 运行成本低,无需新修或改建。但厌氧反应器可能会增加ARGs丰度。与之相比,例如MBR和活性污泥法有较好的处理相关,其中MBR是最有效的技术。但ARGs会在污泥中进行富集,需要继续处理 | [ |
| 消毒工艺 | 消毒工艺具体有紫外消毒、液氯消毒和臭氧消毒等。其中紫外消毒和液氯消毒都可有效降低ARGs丰度,但臭氧消毒只能暂时灭活ARGs,随后促进ARGs的释放 | [ |
| 高级氧化 | 成本较高,产生的羟基自由基(·OH)及光催化中的光辐射损失可使ARB失活,从而对ARGs进行控制和去除 | [ |
| 人工湿地 | 运行成本低、便于维护。去除ARGs的机制为生物降解、底物吸附和植物吸收,在去除ARGs方面有巨大潜力 | [ |
| 组合工艺 | 将不同工艺相组合,优势互补,以达到对ARGs最好的去除效果 | [ |
| 工艺 | 特点 | 参考文献 |
|---|---|---|
| 生物处理 | 运行成本低,无需新修或改建。但厌氧反应器可能会增加ARGs丰度。与之相比,例如MBR和活性污泥法有较好的处理相关,其中MBR是最有效的技术。但ARGs会在污泥中进行富集,需要继续处理 | [ |
| 消毒工艺 | 消毒工艺具体有紫外消毒、液氯消毒和臭氧消毒等。其中紫外消毒和液氯消毒都可有效降低ARGs丰度,但臭氧消毒只能暂时灭活ARGs,随后促进ARGs的释放 | [ |
| 高级氧化 | 成本较高,产生的羟基自由基(·OH)及光催化中的光辐射损失可使ARB失活,从而对ARGs进行控制和去除 | [ |
| 人工湿地 | 运行成本低、便于维护。去除ARGs的机制为生物降解、底物吸附和植物吸收,在去除ARGs方面有巨大潜力 | [ |
| 组合工艺 | 将不同工艺相组合,优势互补,以达到对ARGs最好的去除效果 | [ |
| 技术 | 工艺 | 污水 | 效果 | 参考文献 |
|---|---|---|---|---|
| 生物处理 | MBR | 猪场废水 | ARGs拷贝数去除率为1.5~2 logs | [ |
| SMBR | 较短的SRT下,ARGs的去除率最高,减少了2.91 logs | [ | ||
| CAS | 生活污水 | ARB浓度下降1 logs | [ | |
| 凝灰岩连续种植人工湿地 | int1和总ARGs去除率分别能达到85.9%~89.4%和87.1%~97% | [ | ||
| 人工湿地 | ARGs减少了1~3个数量级 | [ | ||
| BAF | ARGs减少了0.6~1.2个数量级 | |||
| 消毒工艺 | 紫外UV254 | 污水处理厂二沉池和出水池废水 | 5个tet去除率分别在52.0%~73.5%之间(低通量,40mJ/cm2)和79.7%~92.0%之间(高通量,160mJ/cm2) | [ |
| 次氯酸钠(NaClO) | 在8mg Cl2/L甚至更低的情况下,tet去除率可达85%。在16mg Cl2/L,sul去除率可达99% | |||
| O3 | 2mg/L O3下,tet和sul平均去除率为49.2%和34.5% | |||
| 氯化 | 自然水体 | sulI、sulII、tetG和tetA的去除率超过98.0% | [ | |
| 高级氧化 | 石墨氮化碳(g-C3N4)光催化 | 生活污水 | tetA和blaTEM-1的去除率分别为99.0%和98.0% | [ |
| 硫酸纳米锌铁(S-nZVI)过硫酸盐活化 | 5min内胞外ARGs减少8.0 logs | [ | ||
| 石墨氮化碳(g-C3N4)过硫酸盐活化 | tetA和int1减少超过2.5 logs | [ | ||
| CeO2@CNT-NaClO电催化 | 模拟废水 | 30min内tetA和sul1减少2.3 logs和4.6 logs | [ | |
| 氯氧化铋(BiOCl)光催化 | 医院废水 | ampC、mexB、mexD、blaTEM、blaSHV等去除率超过90% | [ | |
| 组合工艺 | OD+反硝化沉淀池+氯消毒 | 生活污水 | 对总ARGs和总MGEs的去除率分别为28.31%和28.76% | [ |
| CASS+反硝化沉淀池+紫外消毒 | 对总ARGs和总MGEs的去除率分别为36.62%和17.34% | |||
| A2/O+MBR+氯消毒 | 对总ARGs和总MGEs的去除率分别为52.04%和44.27% | |||
| 光催化+人工湿地 | 模拟二级出水 | 总ARGs降低了2.36 logs,e-sul1降低了3.07个数量级,e-intI1降低了0.45个数量级 | [ | |
| A/O-MBR-O3/AC | 生活污水 | ARGs降低2~3个数量级,丰度为10-0.71~101.91 copies/mL | [ | |
| UASB/BTF+紫外 | 生活污水 | ARB浓度下降0.5 logs | [ | |
| MAS/UV | 生活污水 | ARB浓度下降2~3 logs | ||
| 次氯酸钠(NaClO)+Fenton | 生活污水 | ARGs减少1.8~4.17 logs,并有效抑制ARB再生 | [ |
| 技术 | 工艺 | 污水 | 效果 | 参考文献 |
|---|---|---|---|---|
| 生物处理 | MBR | 猪场废水 | ARGs拷贝数去除率为1.5~2 logs | [ |
| SMBR | 较短的SRT下,ARGs的去除率最高,减少了2.91 logs | [ | ||
| CAS | 生活污水 | ARB浓度下降1 logs | [ | |
| 凝灰岩连续种植人工湿地 | int1和总ARGs去除率分别能达到85.9%~89.4%和87.1%~97% | [ | ||
| 人工湿地 | ARGs减少了1~3个数量级 | [ | ||
| BAF | ARGs减少了0.6~1.2个数量级 | |||
| 消毒工艺 | 紫外UV254 | 污水处理厂二沉池和出水池废水 | 5个tet去除率分别在52.0%~73.5%之间(低通量,40mJ/cm2)和79.7%~92.0%之间(高通量,160mJ/cm2) | [ |
| 次氯酸钠(NaClO) | 在8mg Cl2/L甚至更低的情况下,tet去除率可达85%。在16mg Cl2/L,sul去除率可达99% | |||
| O3 | 2mg/L O3下,tet和sul平均去除率为49.2%和34.5% | |||
| 氯化 | 自然水体 | sulI、sulII、tetG和tetA的去除率超过98.0% | [ | |
| 高级氧化 | 石墨氮化碳(g-C3N4)光催化 | 生活污水 | tetA和blaTEM-1的去除率分别为99.0%和98.0% | [ |
| 硫酸纳米锌铁(S-nZVI)过硫酸盐活化 | 5min内胞外ARGs减少8.0 logs | [ | ||
| 石墨氮化碳(g-C3N4)过硫酸盐活化 | tetA和int1减少超过2.5 logs | [ | ||
| CeO2@CNT-NaClO电催化 | 模拟废水 | 30min内tetA和sul1减少2.3 logs和4.6 logs | [ | |
| 氯氧化铋(BiOCl)光催化 | 医院废水 | ampC、mexB、mexD、blaTEM、blaSHV等去除率超过90% | [ | |
| 组合工艺 | OD+反硝化沉淀池+氯消毒 | 生活污水 | 对总ARGs和总MGEs的去除率分别为28.31%和28.76% | [ |
| CASS+反硝化沉淀池+紫外消毒 | 对总ARGs和总MGEs的去除率分别为36.62%和17.34% | |||
| A2/O+MBR+氯消毒 | 对总ARGs和总MGEs的去除率分别为52.04%和44.27% | |||
| 光催化+人工湿地 | 模拟二级出水 | 总ARGs降低了2.36 logs,e-sul1降低了3.07个数量级,e-intI1降低了0.45个数量级 | [ | |
| A/O-MBR-O3/AC | 生活污水 | ARGs降低2~3个数量级,丰度为10-0.71~101.91 copies/mL | [ | |
| UASB/BTF+紫外 | 生活污水 | ARB浓度下降0.5 logs | [ | |
| MAS/UV | 生活污水 | ARB浓度下降2~3 logs | ||
| 次氯酸钠(NaClO)+Fenton | 生活污水 | ARGs减少1.8~4.17 logs,并有效抑制ARB再生 | [ |
| 1 | 李超, 关芳灵, 任建军, 等. 抗生素的历史演进及思考[J]. 医学与哲学(B), 2015, 36(11): 80-83. |
| LI Chao, GUAN Fangling, REN Jianjun, et al. The historical evolution and thinking of antibiotics[J]. Medicine & Philosophy(B), 2015, 36(11): 80-83. | |
| 2 | 刘艳英. 抗生素抗性基因在环境中的分布及对人体的危害[J]. 厦门科技, 2020(6): 31-34. |
| LIU Yanying. Distribution of antibiotic resistance genes in the environment and the risk to humans[J]. Xiamen Science &Technology, 2020(6): 31-34. | |
| 3 | JIAN Zonghui, ZENG Li, XU Taojie, et al. Antibiotic resistance genes in bacteria: Occurrence, spread, and control[J]. Journal of Basic Microbiology, 2021, 61(12): 1049-1070. |
| 4 | 朱永官, 陈青林, 苏建强, 等. 环境中抗生素与抗性基因组的研究[J]. 科学观察, 2017, 12(6): 60-62. |
| ZHU Yongguan, CHEN Qinglin, SU Jianqiang, et al. Study of antibiotics and resistance genomes in the environment[J]. Science Focus, 2017, 12(6): 60-62. | |
| 5 | 朱永官, 欧阳纬莹, 吴楠, 等. 抗生素耐药性的来源与控制对策[J]. 中国科学院院刊, 2015, 30(4): 509-516. |
| ZHU Yongguan, OUYANG Weiying, WU Nan, et al. Antibiotic resistance: Sources and mitigation[J]. Bulletin of Chinese Academy of Sciences, 2015, 30(4): 509-516. | |
| 6 | FORSBERG Kevin J, REYES Alejandro, WANG Bin, et al. The shared antibiotic resistome of soil bacteria and human pathogens[J]. Science, 2012, 337(6098): 1107-1111. |
| 7 | 周谊霞, 田永明. 急危重症护理学[M]. 北京: 中国医药科技出版社, 2016. |
| ZHOU Yixia, TIAN Yongming. Critical care[M]. Beijing: China Medical Science Press, 2016. | |
| 8 | GUIDOS Robert, SPELLBERG Brad, BLASER Martin, et al. Combating antimicrobial resistance: Policy recommendations to save lives[J]. Clinical Infectious Diseases, 2011, 52(S5): S397-S428. |
| 9 | KEELY S P, BRINKMAN N E, WHEATON E A, et al. Geospatial patterns of antimicrobial resistance genes in the US EPA national rivers and streams assessment survey[J]. Environmental Science & Technology, 2022, 56(21): 14960-14971. |
| 10 | MACHADO Elayne Cristina, FREITAS Deborah Leroy, LEAL Cintia Dutra, et al. Antibiotic resistance profile of wastewater treatment plants in Brazil reveals different patterns of resistance and multi resistant bacteria in final effluents[J]. Science of the Total Environment, 2023, 857: 159376. |
| 11 | CHEN Qinglin, AN Xinli, LI Hu, et al. Long-term field application of sewage sludge increases the abundance of antibiotic resistance genes in soil[J]. Environment International, 2016, 92: 1-10. |
| 12 | SU Jianqiang, WEI Bei, OUYANG Weiying, et al. Antibiotic resistome and its association with bacterial communities during sewage sludge composting[J]. Environmental Science & Technology, 2015, 49(12): 7356-7363. |
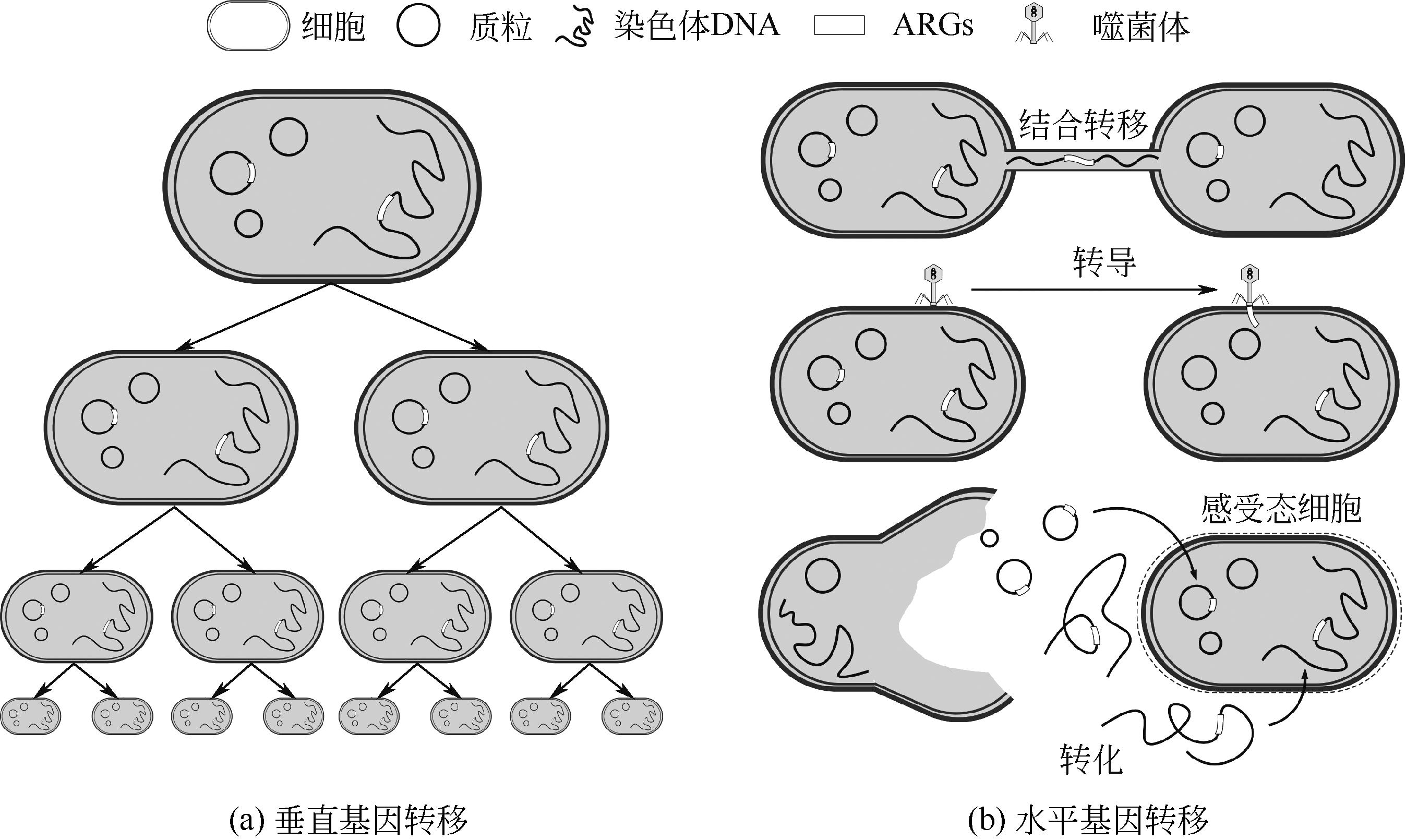
| 13 | SOUCY Shannon M, HUANG Jinling, GOGARTEN Johann Peter. Horizontal gene transfer: Building the web of life[J]. Nature Reviews Genetics, 2015, 16(8): 472-482. |
| 14 | SHAO Sicheng, HU Yongyou, CHENG Jianhua, et al. Research progress on distribution, migration, transformation of antibiotics and antibiotic resistance genes (ARGs) in aquatic environment[J]. Critical Reviews in Biotechnology, 2018, 38(8): 1195-1208. |
| 15 | OCHMAN H, LAWRENCE J G, GROISMAN E A. Lateral gene transfer and the nature of bacterial innovation[J]. Nature, 2000, 405(6784): 299-304. |
| 16 | 周宏瑞, 杨雨桐, 杨晓波, 等. 纳米二硫化钼促进粪肠球菌中信息素诱导质粒介导的耐药基因接合转移[J]. 生态毒理学报, 2022, 17(1): 160-169. |
| ZHOU Hongrui, YANG Yutong, YANG Xiaobo, et al. Molybdenum disulfide promotes pheromone-induced plasmid mediated conjugation transfer of drug resistance genes in enterococcus faecalis[J]. Asian Journal of Ecotoxicology, 2022, 17(1): 160-169. | |
| 17 | LU Ji, WANG Yue, JIN Min, et al. Both silver ions and silver nanoparticles facilitate the horizontal transfer of plasmid-mediated antibiotic resistance genes[J]. Water Research, 2020, 169: 115229. |
| 18 | LI Ping, LUO Wanying, XIANG Tianxin, et al. Horizontal gene transfer via OMVs co-carrying virulence and antimicrobial-resistant genes is a novel way for the dissemination of carbapenem-resistant hypervirulent Klebsiella pneumoniae [J]. Frontiers in Microbiology, 2022, 13: 945972. |
| 19 | LI Bing, QIU Yong, SONG Yanqing, et al. Dissecting horizontal and vertical gene transfer of antibiotic resistance plasmid in bacterial community using microfluidics[J]. Environment International, 2019, 131: 105007. |
| 20 | MUNIESA Maite, Marta COLOMER-LLUCH, JOFRE Juan. Potential impact of environmental bacteriophages in spreading antibiotic resistance genes[J]. Future Microbiology, 2013, 8(6): 739-751. |
| 21 | 张越, 胡雪莹, 王旭明. 噬菌体编码的抗生素抗性基因研究进展[J]. 中国环境科学, 2022, 42(5): 2315-2320. |
| ZHANG Yue, HU Xueying, WANG Xuming. Recent advances on antibiotic resistance genes encoded by bacteriophages[J]. China Environmental Science, 2022, 42(5): 2315-2320. | |
| 22 | KENZAKA Takehiko, TANI Katsuji, NASU Masao. High-frequency phage-mediated gene transfer in freshwater environments determined at single-cell level[J]. The ISME Journal, 2010, 4(5): 648-659. |
| 23 | BAHAROGLU Zeynep, MAZEL Didier. Vibrio cholerae triggers SOS and mutagenesis in response to a wide range of antibiotics: A route towards multiresistance[J]. Antimicrobial Agents and Chemotherapy, 2011, 55(5): 2438-2441. |
| 24 | Uli KLÜMPER, RIBER Leise, DECHESNE Arnaud, et al. Broad host range plasmids can invade an unexpectedly diverse fraction of a soil bacterial community[J]. The ISME Journal, 2015, 9(4): 934-945. |
| 25 | Søren OVERBALLE-PETERSEN, HARMS Klaus, ORLANDO Ludovic A A, et al. Bacterial natural transformation by highly fragmented and damaged DNA[J]. Proceedings of the National Academy of Sciences of the United States of America, 2013, 110(49): 19860-19865. |
| 26 | CHEN Inês, DUBNAU David. DNA uptake during bacterial transformation[J]. Nature Reviews Microbiology, 2004, 2(3): 241-249. |
| 27 | 李文化. Ca2+诱导大肠杆菌摄取外源DNA的研究[D]. 武汉: 武汉大学, 2004. |
| LI Wenhua. Ca2+-induced uptake of foreign DNA by Escherichia coli [D]. Wuhan: Wuhan University, 2004. | |
| 28 | DING Chengshi, PAN Jie, JIN Min, et al. Enhanced uptake of antibiotic resistance genes in the presence of nanoalumina[J]. Nanotoxicology, 2016, 10(8): 1051-1060. |
| 29 | WANG Yue, LU Ji, Jan ENGELSTÄDTER, et al. Non-antibiotic pharmaceuticals enhance the transmission of exogenous antibiotic resistance genes through bacterial transformation[J]. The ISME Journal, 2020, 14(8): 2179-2196. |
| 30 | BASU Shreya, MUKHERJEE Mandira. Incidence and risk of co-transmission of plasmid-mediated quinolone resistance and extended-spectrum β - l a c t a m a s e genes in fluoroquinolone-resistant uropathogenic Escherichia coli: A first study from Kolkata, India[J]. Journal of Global Antimicrobial Resistance, 2018, 14: 217-223. |
| 31 | LI Dong, YU Tao, ZHANG Yu, et al. Antibiotic resistance characteristics of environmental bacteria from an oxytetracycline production wastewater treatment plant and the receiving river[J]. Applied and Environmental Microbiology, 2010, 76(11): 3444-3451. |
| 32 | ZHAO Renxin, FENG Jie, LIU Jie, et al. Deciphering of microbial community and antibiotic resistance genes in activated sludge reactors under high selective pressure of different antibiotics[J]. Water Research, 2019, 151: 388-402. |
| 33 | ZHAO Qi, GUO Wanqian, LUO Haichao, et al. Deciphering the transfers of antibiotic resistance genes under antibiotic exposure conditions: Driven by functional modules and bacterial community[J]. Water Research, 2021, 205: 117672. |
| 34 | ZHAO Renxin, YU Ke, ZHANG Jiayu, et al. Deciphering the mobility and bacterial hosts of antibiotic resistance genes under antibiotic selection pressure by metagenomic assembly and binning approaches[J]. Water Research, 2020, 186: 116318. |
| 35 | 徐立冬, 董莉倩, 粱秀峰, 等. 抗生素压力下铜绿假单胞菌耐药基因动态监测分析[J]. 中国卫生检验杂志, 2023, 33(16): 1946-1948. |
| XU Lidong, DONG Liqian, LIANG Xiufeng, et al. Dynamic monitoring and analysis of resistant gene of Pseudomonas aeruginosa under antibiotic pressure[J]. Chinese Journal of Health Laboratory Technology, 2023, 33(16): 1946-1948. | |
| 36 | GULLBERG Erik, ALBRECHT Lisa M, KARLSSON Christoffer, et al. Selection of a multidrug resistance plasmid by sublethal levels of antibiotics and heavy metals[J]. mBio, 2014, 5(5): e01918-14. |
| 37 | SONG Jianxiao, RENSING Christopher, HOLM Peter E, et al. Comparison of metals and tetracycline as selective agents for development of tetracycline resistant bacterial communities in agricultural soil[J]. Environmental Science & Technology, 2017, 51(5): 3040-3047. |
| 38 | GUPTA Sonia, GRAHAM David W, SREEKRISHNAN T R, et al. Heavy metal and antibiotic resistance in four Indian and UK rivers with different levels and types of water pollution[J]. Science of the Total Environment, 2023, 857: 159059. |
| 39 | WANG Xiaomin, LAN Bangrui, FEI Hexin, et al. Heavy metal could drive co-selection of antibiotic resistance in terrestrial subsurface soils[J]. Journal of Hazardous Materials, 2021, 411: 124848. |
| 40 | XIA Juntao, SUN Haohao, ZHANG Xuxiang, et al. Aromatic compounds lead to increased abundance of antibiotic resistance genes in wastewater treatment bioreactors[J]. Water Research, 2019, 166: 115073. |
| 41 | YU Kaiqiang, CHEN Feiran, YUE Le, et al. CeO2 nanoparticles regulate the propagation of antibiotic resistance genes by altering cellular contact and plasmid transfer[J]. Environmental Science & Technology, 2020, 54(16): 10012-10021. |
| 42 | CHEN Qinglin, ZHU Dong, AN Xinli, et al. Does nano silver promote the selection of antibiotic resistance genes in soil and plant?[J]. Environment International, 2019, 128: 399-406. |
| 43 | 董天羽. 城市河流抗生素抗性基因污染特征及转化研究[D]. 邯郸: 河北工程大学, 2022. |
| DONG Tianyu. Study on pollution characteristics and transformation of antibiotic resistance genes in urban rivers[D]. Handan: Hebei University of Engineering, 2022. | |
| 44 | HAN Xue, Peng LYU, WANG Luguang, et al. Impact of nano-TiO2 on horizontal transfer of resistance genes mediated by filamentous phage transduction[J]. Environmental Science: Nano, 2020, 7(4): 1214-1224. |
| 45 | XIAO Xiang, MA Xiaolin, HAN Xue, et al. TiO2 photoexcitation promoted horizontal transfer of resistance genes mediated by phage transduction[J]. Science of the Total Environment, 2021, 760: 144040. |
| 46 | SEILER Claudia, BERENDONK Thomas U. Heavy metal driven co-selection of antibiotic resistance in soil and water bodies impacted by agriculture and aquaculture[J]. Frontiers in Microbiology, 2012, 3: 399. |
| 47 | GHADERPOUR Aziz, Wing Sze HO, CHEW Li-Lee, et al. Diverse and abundant multi-drug resistant E.coli in Matang mangrove estuaries, Malaysia[J]. Frontiers in Microbiology, 2015, 6: 977. |
| 48 | ENGIN Ayse Basak, ENGIN Evren Doruk, ENGIN Atilla. Effects of co-selection of antibiotic-resistance and metal-resistance genes on antibiotic-resistance potency of environmental bacteria and related ecological risk factors[J]. Environmental Toxicology and Pharmacology, 2023, 98: 104081. |
| 49 | 薛喜枚, 朱永官. 土壤中砷的生物转化及砷与抗生素抗性的关联[J]. 土壤学报, 2019, 56(4): 763-772. |
| XUE Ximei, ZHU Yongguan. Arsenic biotransformation in soils and its relationship with antibiotic resistance[J]. Acta Pedologica Sinica, 2019, 56(4): 763-772. | |
| 50 | 林燕. 抗生素大环境下的保健养殖如何开展?[J]. 猪业科学, 2012, 29(9): 98. |
| LIN Yan. How to carry out health-care farming under the environment of antibiotics?[J]. Swine Industry Science, 2012, 29(9): 98. | |
| 51 | 李蕊宁. 改性马铃薯秸秆生物炭对水体中典型抗生素的吸附性能研究[D]. 兰州: 兰州大学, 2018. |
| LI Ruining. Study on adsorption performance of modified potato straw biochar for typical antibiotics in water[D]. Lanzhou: Lanzhou University, 2018. | |
| 52 | 王晓慧. 抗生素工业废水厌氧生物处理技术研究[D]. 郑州: 郑州大学, 2005. |
| WANG Xiaohui. Study on anaerobic biological treatment technology of antibiotic industrial wastewater[D]. Zhengzhou: Zhengzhou University, 2005. | |
| 53 | 齐亚兵, 张思敬, 孟晓荣, 等. 抗生素废水处理技术现状及研究进展[J]. 应用化工, 2021, 50(9): 2587-2593, 2597. |
| QI Yabing, ZHANG Sijing, MENG Xiaorong, et al. The present situation and research progress of antibiotic wastewater treatment technology[J]. Applied Chemical Industry, 2021, 50(9): 2587-2593, 2597. | |
| 54 | 洪颖忻, 吴浪, 张立秋, 等. 制药废水处理系统中抗生素抗性基因的研究进展[J]. 工业水处理, 2022, 42(4): 39-45. |
| HONG Yingxin, WU Lang, ZHANG Liqiu, et al. Research progress of antibiotic resistance genes in pharmaceutical wastewater treatment systems[J]. Industrial Water Treatment, 2022, 42(4): 39-45. | |
| 55 | 刘影, 张晋东, 吴艳丽, 等. 畜牧业抗生素抗性基因的分布与传播研究进展[J]. 家畜生态学报, 2024, 45(4): 1-7. |
| LIU Ying, ZHANG Jindong, WU Yanli, et al. Advances in the distribution and dissemination of antibiotic resistance genes in animal husbandry[J]. Journal of Domestic Animal Ecology, 2024, 45(4): 1-7. | |
| 56 | 凌文翠, 范玉梅, 方瑶瑶, 等. 京津冀地区畜禽养殖业抗生素污染现状分析[J]. 环境工程技术学报, 2018, 8(4): 390-397. |
| LING Wencui, FAN Yumei, FANG Yaoyao, et al. Antibiotics pollution of livestock and poultry breeding in Beijing-Tianjin-Hebei region[J]. Journal of Environmental Engineering Technology, 2018, 8(4): 390-397. | |
| 57 | 姚小文. 铁碳微电解-Fenton+两级A/O工艺处理抗生素废水的应用研究[D]. 南昌: 南昌大学, 2019. |
| YAO Xiaowen. Study on the application of iron-carbon micro-electrolysis-Fenton+ two-stage A/O process to treat antibiotic wastewater[D]. Nanchang: Nanchang University, 2019. | |
| 58 | 徐国韬, 周晓琴, 李子富, 等. 尿液中药物与耐药污染及削减技术研究进展[J]. 中国环境科学, 2024, 44(2): 1122-1133. |
| XU Guotao, ZHOU Xiaoqin, LI Zifu, et al. Progresses on characteristics and reduction technologies of drugs and related drug-resistant contamination in urine[J]. China Environmental Science, 2024, 44(2): 1122-1133. | |
| 59 | 黄圣琳, 何势, 魏欣, 等. 污水处理厂中四环素类抗生素残留及其抗性基因污染特征研究进展[J]. 化工进展, 2015, 34(6): 1779-1785. |
| HUANG Shenglin, HE Shi, WEI Xin, et al. Pollution characteristics of tetracycline residues and tetracycline resistance genes in sewage treatment plants: A review[J]. Chemical Industry and Engineering Progress, 2015, 34(6): 1779-1785. | |
| 60 | DAVIES Julian, DAVIES Dorothy. Origins and evolution of antibiotic resistance[J]. Microbiology and Molecular Biology Reviews, 2010, 74(3): 417-433. |
| 61 | 许文锋, 刘丽华, 黄福义, 等. 厦门市近岸海域水环境抗生素抗性基因污染分布特征[J]. 环境科学, 2024, 45(7): 141-148. |
| XU Wenfeng, LIU Lihua, HUANG Fuyi, et al. Characteristics of the distribution of antibiotic resistance genes in Xiamen's near-shore waters[J]. Environmental Science, 2024, 45(7): 141-148. | |
| 62 | DANG Chenyuan, XIA Yu, ZHENG Maosheng, et al. Metagenomic insights into the profile of antibiotic resistomes in a large drinking water reservoir[J]. Environment International, 2020, 136: 105449. |
| 63 | 王秋水, 程波, 刘悦, 等. 昌黎县海域细菌群落和抗生素抗性基因分析[J]. 环境科学, 2024, 45(1): 567-575. |
| WANG Qiushui, CHENG Bo, LIU Yue, et al. Analysis of bacterial communities and antibiotic resistance genes in the aquaculture area of Changli County[J]. Environmental Science, 2024, 45(1): 567-575. | |
| 64 | 胡一凯. 秦皇岛典型滨海浴场水体中抗生素抗性基因的分布特征及驱动机制[D]. 秦皇岛: 燕山大学, 2023. |
| HU Yikai. Distribution characteristics and driving mechanism of antibiotic resistance genes in typical coastal bathing waters of Qinhuangdao[D]. Qinhuangdao: Yanshan University, 2023. | |
| 65 | YANG Yuyi, LIU Guihua, SONG Wenjuan, et al. Plastics in the marine environment are reservoirs for antibiotic and metal resistance genes[J]. Environment International, 2019, 123: 79-86. |
| 66 | ZHANG Yuxuan, WANG Jianhua, LU Jian, et al. Antibiotic resistance genes might serve as new indicators for wastewater contamination of coastal waters: Spatial distribution and source apportionment of antibiotic resistance genes in a coastal bay[J]. Ecological Indicators, 2020, 114: 106299. |
| 67 | 臧泉. 武汉与西藏湖泊微生物群落多样性及抗生素抗性基因分布特征[D]. 拉萨: 西藏大学, 2021. |
| ZANG Quan. Diversity of microbial community and distribution characteristics of antibiotic resistance genes in lakes of Wuhan and Tibet[D]. Lasa: Tibet University, 2021. | |
| 68 | 吴韵斐, 何义亮, 袁其懿, 等. 水源型水库抗生素抗性基因赋存特征研究[J]. 环境科学学报, 2019, 39(6): 1834-1841. |
| WU Yunfei, HE Yiliang, YUAN Qiyi, et al. Study on antibiotic resistance genes characteristics in a drinking water reservoir[J]. Acta Scientiae Circumstantiae, 2019, 39(6): 1834-1841. | |
| 69 | ZHANG Yongpeng, SHEN Genxiang, HU Shuangqing, et al. Deciphering of antibiotic resistance genes (ARGs) and potential abiotic indicators for the emergence of ARGs in an interconnected lake-river-reservoir system[J]. Journal of Hazardous Materials, 2021, 410: 124552. |
| 70 | SHI Xiaomin, SHEN Zhangqi, SHAO Bing, et al. Antibiotic resistance genes profile in the surface sediments of typical aquaculture areas across 15 major lakes in China[J]. Environmental Pollution, 2024, 347: 123709. |
| 71 | 吴天宇. 赤水河流域水体抗生素抗性基因赋存特征及来源解析[D]. 贵阳: 贵州大学, 2022. |
| WU Tianyu. Occurrence characteristics and source analysis of antibiotic resistance genes in Chishui River basin[D]. Guiyang: Guizhou University, 2022. | |
| 72 | MITTAL Parul, Vishnu Prasoodanan PK, DHAKAN Darshan B, et al. Metagenome of a polluted river reveals a reservoir of metabolic and antibiotic resistance genes[J]. Environmental Microbiome, 2019, 14(1): 5. |
| 73 | ZHANG Guodong, LU Shaoyong, WANG Yongqiang, et al. Occurrence of antibiotics and antibiotic resistance genes and their correlations in Lower Yangtze River, China[J]. Environmental Pollution, 2020, 257: 113365. |
| 74 | ZHANG Lili, ZHANG Cheng, LIAN Keting, et al. River restoration changes distributions of antibiotics, antibiotic resistance genes, and microbial community[J]. Science of the Total Environment, 2021, 788: 147873. |
| 75 | SZEKERES Edina, CHIRIAC Cecilia Maria, BARICZ Andreea, et al. Investigating antibiotics, antibiotic resistance genes, and microbial contaminants in groundwater in relation to the proximity of urban areas[J]. Environmental Pollution, 2018, 236: 734-744. |
| 76 | WU Dailing, ZHANG Min, HE Luxi, et al. Contamination profile of antibiotic resistance genes in ground water in comparison with surface water[J]. Science of the Total Environment, 2020, 715: 136975. |
| 77 | KNAPP Charles W, ZHANG Wen, STURM Belinda S M, et al. Differential fate of erythromycin and beta-lactam resistance genes from swine lagoon waste under different aquatic conditions[J]. Environmental Pollution, 2010, 158(5): 1506-1512. |
| 78 | MINDLIN Sofia, MINAKHIN Leonid, PETROVA Mayya, et al. Present-day mercury resistance transposons are common in bacteria preserved in permafrost grounds since the Upper Pleistocene[J]. Research in Microbiology, 2005, 156(10): 994-1004. |
| 79 | Zachary CHARLOP-POWERS, OWEN Jeremy G, REDDY Boojala Vijay B, et al. Global biogeographic sampling of bacterial secondary metabolism[J]. eLife, 2015, 4: e05048. |
| 80 | MARTÍNEZ José L. Antibiotics and antibiotic resistance genes in natural environments[J]. Science, 2008, 321(5887): 365-367. |
| 81 | 赵方杰, 谢婉滢, 汪鹏. 土壤与人体健康[J]. 土壤学报, 2020, 57(1): 1-11. |
| ZHAO Fangjie, XIE Wanying, WANG Peng. Soil and human health[J]. Acta Pedologica Sinica, 2020, 57(1): 1-11. | |
| 82 | CASTANON J I R. History of the use of antibiotic as growth promoters in European poultry feeds[J]. Poultry Science, 2007, 86(11): 2466-2471. |
| 83 | 武晨, 黄凤莲, 刘新刚, 等. 环洞庭湖土壤抗生素抗性基因分布和潜在风险[J]. 中国环境科学, 2024, 44(3): 1575-1583. |
| WU Chen, HUANG Fenglian, LIU Xingang, et al. Distributions and potential risks of antibiotic resistance genes in soils around Dongting Lake Basin, China[J]. China Environmental Science, 2024, 44(3): 1575-1583. | |
| 84 | ZHU Yongguan, JOHNSON Timothy A, SU Jianqiang, et al. Diverse and abundant antibiotic resistance genes in Chinese swine farms[J]. Proceedings of the National Academy of Sciences of the United States of America, 2013, 110(9): 3435-3440. |
| 85 | XIE Wanying, YUAN Shuangting, XU Minggang, et al. Long-term effects of manure and chemical fertilizers on soil antibiotic resistome[J]. Soil Biology and Biochemistry, 2018, 122: 111-119. |
| 86 | YU Zhongyi, GUNN Lynda, WALL Patrick, et al. Antimicrobial resistance and its association with tolerance to heavy metals in agriculture production[J]. Food Microbiology, 2017, 64: 23-32. |
| 87 | PAZDA Magdalena, KUMIRSKA Jolanta, STEPNOWSKI Piotr, et al. Antibiotic resistance genes identified in wastewater treatment plant systems—A review[J]. Science of the Total Environment, 2019, 697: 134023. |
| 88 | WANG Jianlong, CHU Libing, László WOJNÁROVITS, et al. Occurrence and fate of antibiotics, antibiotic resistant genes (ARGs) and antibiotic resistant bacteria (ARB) in municipal wastewater treatment plant: An overview[J]. Science of the Total Environment, 2020, 744: 140997. |
| 89 | NGUYEN Anh Q, Hang P VU, NGUYEN Luong N, et al. Monitoring antibiotic resistance genes in wastewater treatment: Current strategies and future challenges[J]. Science of the Total Environment, 2021, 783: 146964. |
| 90 | ZHU Tingting, SU Zhongxian, LAI Wenxia, et al. Insights into the fate and removal of antibiotics and antibiotic resistance genes using biological wastewater treatment technology[J]. Science of the Total Environment, 2021, 776: 145906. |
| 91 | EZEUKO Adaora S, OJEMAYE Mike O, OKOH Omobola O, et al. Technological advancement for eliminating antibiotic resistance genes from wastewater: A review of their mechanisms and progress[J]. Journal of Environmental Chemical Engineering, 2021, 9(5): 106183. |
| 92 | PHUONG HOA Phan Thi, NONAKA Lisa, HUNG VIET Pham, et al. Detection of the Sul1, Sul2, and Sul3 genes in sulfonamide-resistant bacteria from wastewater and shrimp ponds of north Vietnam[J]. Science of the Total Environment, 2008, 405(1/2/3): 377-384. |
| 93 | FAN Xiaoyan, GAO Jingfeng, PAN Kailing, et al. Functional Genera, potential pathogens and predicted antibiotic resistance genes in 16 full-scale wastewater treatment plants treating different types of wastewater[J]. Bioresource Technology, 2018, 268: 97-106. |
| 94 | CHRISTGEN Beate, YANG Ying, AHAMMAD S Z, et al. Metagenomics shows that low-energy anaerobic-aerobic treatment reactors reduce antibiotic resistance gene levels from domestic wastewater[J]. Environmental Science & Technology, 2015, 49(4): 2577-2584. |
| 95 | 段彤, 曾小芸, 谈树成. MBR处理猪场废水过程中抗生素抗性基因的去除[J]. 环境工程, 2022, 40(4): 8-13, 21. |
| DUAN Tong, ZENG Xiaoyun, TAN Shucheng. Removal of antibiotic resistance genes during the treatment of swine wastewater by MBR[J]. Environmental Engineering, 2022, 40(4): 8-13, 21. | |
| 96 | LI Bing, QIU Yong, LI Ji, et al. Removal of antibiotic resistance genes in four full-scale membrane bioreactors[J]. Science of the Total Environment, 2019, 653: 112-119. |
| 97 | MUNIR Mariya, WONG Kelvin, XAGORARAKI Irene. Release of antibiotic resistant bacteria and genes in the effluent and biosolids of five wastewater utilities in Michigan[J]. Water Research, 2011, 45(2): 681-693. |
| 98 | LE Thai-Hoang, Charmaine NG, TRAN Ngoc Han, et al. Removal of antibiotic residues, antibiotic resistant bacteria and antibiotic resistance genes in municipal wastewater by membrane bioreactor systems[J]. Water Research, 2018, 145: 498-508. |
| 99 | STANGE C, SIDHU J P S, TOZE S, et al. Comparative removal of antibiotic resistance genes during chlorination, ozonation, and UV treatment[J]. International Journal of Hygiene and Environmental Health, 2019, 222(3): 541-548. |
| 100 | ZHENG Ji, SU Chao, ZHOU Jianwen, et al. Effects and mechanisms of ultraviolet, chlorination, and ozone disinfection on antibiotic resistance genes in secondary effluents of municipal wastewater treatment plants[J]. Chemical Engineering Journal, 2017, 317: 309-316. |
| 101 | 吴文瞳, 张玲玲, 李子富, 等. 高级氧化技术降解抗生素及去除耐药性的研究进展[J]. 化工进展, 2021, 40(8): 4551-4561. |
| WU Wentong, ZHANG Lingling, LI Zifu, et al. Research progress of advanced oxidation technology in degradation of antibiotics and removal of antibiotic resistance[J]. Chemical Industry and Engineering Progress, 2021, 40(8): 4551-4561. | |
| 102 | LIU Xiaohui, GUO Xiaochun, LIU Ying, et al. A review on removing antibiotics and antibiotic resistance genes from wastewater by constructed wetlands: Performance and microbial response[J]. Environmental Pollution, 2019, 254: 112996. |
| 103 | CHEN Jun, YING Guangguo, WEI Xiaodong, et al. Removal of antibiotics and antibiotic resistance genes from domestic sewage by constructed wetlands: Effect of flow configuration and plant species[J]. Science of the Total Environment, 2016, 571: 974-982. |
| 104 | HUANG Xu, LIU Chaoxiang, LI Ke, et al. Performance of vertical up-flow constructed wetlands on swine wastewater containing tetracyclines and Tet genes[J]. Water Research, 2015, 70: 109-117. |
| 105 | 王子木. 不同污水处理对污水处理厂废水中抗生素抗性基因和抗性菌的去除效果[D]. 雅安: 四川农业大学, 2023. |
| WANG Zimu. Removal effect of different sewage treatments on antibiotic resistance genes and resistant bacteria in wastewater from sewage treatment plants[D]. Ya'an: Sichuan Agricultural University, 2023. | |
| 106 | SUI Qianwen, JIANG Chao, ZHANG Junya, et al. Does the biological treatment or membrane separation reduce the antibiotic resistance genes from swine wastewater through a sequencing-batch membrane bioreactor treatment process[J]. Environment International, 2018, 118: 274-281. |
| 107 | Ammar ABOU-KANDIL, SHIBLI Areen, AZAIZEH Hassan, et al. Fate and removal of bacteria and antibiotic resistance genes in horizontal subsurface constructed wetlands: Effect of mixed vegetation and substrate type[J]. Science of the Total Environment, 2021, 759: 144193. |
| 108 | CHEN Hong, ZHANG Mingmei. Effects of advanced treatment systems on the removal of antibiotic resistance genes in wastewater treatment plants from Hangzhou, China[J]. Environmental Science & Technology, 2013, 47(15): 8157-8163. |
| 109 | GUO Xueping, LI Jing, YANG Fan, et al. Prevalence of sulfonamide and tetracycline resistance genes in drinking water treatment plants in the Yangtze River Delta, China[J]. Science of the Total Environment, 2014, 493: 626-631. |
| 110 | ZHONG Jiexi, AHMED Yunus, CARVALHO Gilda, et al. Simultaneous removal of micropollutants, antibiotic resistant bacteria, and antibiotic resistance genes using graphitic carbon nitride under simulated solar irradiation[J]. Chemical Engineering Journal, 2022, 433: 133839. |
| 111 | YU Zhigang, RABIEE Hesamoddin, GUO Jianhua. Synergistic effect of sulfidated nano zerovalent iron and persulfate on inactivating antibiotic resistant bacteria and antibiotic resistance genes[J]. Water Research, 2021, 198: 117141. |
| 112 | GAO Boru, DOU Mengmeng, WANG Jin, et al. Efficient persulfate activation by carbon defects g-C3N4 containing electron traps for the removal of antibiotics, resistant bacteria and genes[J]. Chemical Engineering Journal, 2021, 426: 131677. |
| 113 | NI Xiaoyu, HOU Xuan, MA Defang, et al. Simultaneous removal of antibiotics and antibiotic resistant genes using a CeO2@CNT electrochemical membrane-NaClO system[J]. Chemosphere, 2023, 338: 139457. |
| 114 | LIU Mingyang, ZHU Hangqi, ZHU Nali, et al. Vacancy engineering of BiOCl microspheres for efficient removal of multidrug-resistant bacteria and antibiotic-resistant genes in wastewater[J]. Chemical Engineering Journal, 2021, 426: 130710. |
| 115 | 谌萍萍. 光催化-人工湿地去除抗生素抗性基因技术及其机制[D]. 长春: 东北师范大学, 2023. |
| CHEN Pingping. Technology and mechanism of removing antibiotic resistance genes by photocatalysis-constructed wetland[D]. Changchun: Northeast Normal University, 2023. | |
| 116 | 冯萃敏, 庆杉, 安鑫悦, 等. MBR工艺对公共建筑污水抗生素抗性基因的去除效果研究[J]. 安全与环境学报, 2020, 20(2): 683-689. |
| FENG Cuimin, QING Shan, AN Xinyue, et al. Removal efficiency of antibiotic resistance genes in sewage from public buildings by membrane bioreactor[J]. Journal of Safety and Environment, 2020, 20(2): 683-689. | |
| 117 | LUO Ling, WANG Zimu, HUANG Xin, et al. The fate of antibiotic resistance genes in wastewater containing microalgae treated by chlorination, ultra-violet, and Fenton reaction[J]. Water Research, 2024, 254: 121392. |
| 118 | WU Yinhu, WANG Yunhong, XUE Song, et al. Increased risks of antibiotic resistant genes (ARGs) induced by chlorine disinfection in the reverse osmosis system for potable reuse of reclaimed water[J]. Science of the Total Environment, 2022, 815: 152860. |
| 119 | LI Hu, SONG Ruiying, WANG Yangyang, et al. Inhibited conjugative transfer of antibiotic resistance genes in antibiotic resistant bacteria by surface plasma[J]. Water Research, 2021, 204: 117630. |
| [1] | TANG Jiaojiao, XIE Junxiang, CHEN Chongjun, YU Cheng, CHEN Dechao. Carbon neutral technologies and case studies in urban sewage treatment plants [J]. Chemical Industry and Engineering Progress, 2022, 41(5): 2662-2671. |
| [2] | ZHANG Huining, SHI Zhongyu, XIAO Yankui, ZHANG Xiaoqin, YIN Xin, TIAN Lihong. Preparation of 3D graphene by 3D printing and its application in water treatment [J]. Chemical Industry and Engineering Progress, 2022, 41(5): 2231-2242. |
| [3] | LI Dongmei, WU Danping, WU Min, PAN Bo. Influence of operating parameters on greenhouse gas emission of sewage treatment plants [J]. Chemical Industry and Engineering Progress, 2021, 40(12): 6897-6906. |
| [4] | HUANG Shenglin, HE Shi, WEI Xin, XUE Gang, GAO Pin. Pollution characteristics of tetracycline residues and tetracycline resistance genes in sewage treatment plants:A review [J]. Chemical Industry and Engineering Progree, 2015, 34(06): 1779-1785. |
| [5] | CHENG Chen, YAN Zheng. Degradation of oxytetracycline in wastewater by multi-frequency ultrasonic [J]. Chemical Industry and Engineering Progree, 2015, 34(04): 1143-1146,1164. |
| [6] | ZHANG Fan,LI Jing,TAN Jianhua,WANG Bo,HUANG Fu. Advance of the treatment of heavy metal wastewater by adsorption [J]. Chemical Industry and Engineering Progree, 2013, 32(11): 2749-2756. |
| [7] | YANG Li1,LIAO Chuanhua2,ZHU Yuezhao2, CHEN Haijun2,JIN Qinfang1. Characteristics of micro-bubble and nano-bubble and their application in environmental pollution control [J]. Chemical Industry and Engineering Progree, 2012, 31(06): 1333-1337. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||